Fig. 1. Schematic of cerebral organoid and TMT-LC/MS analytical pipeline.
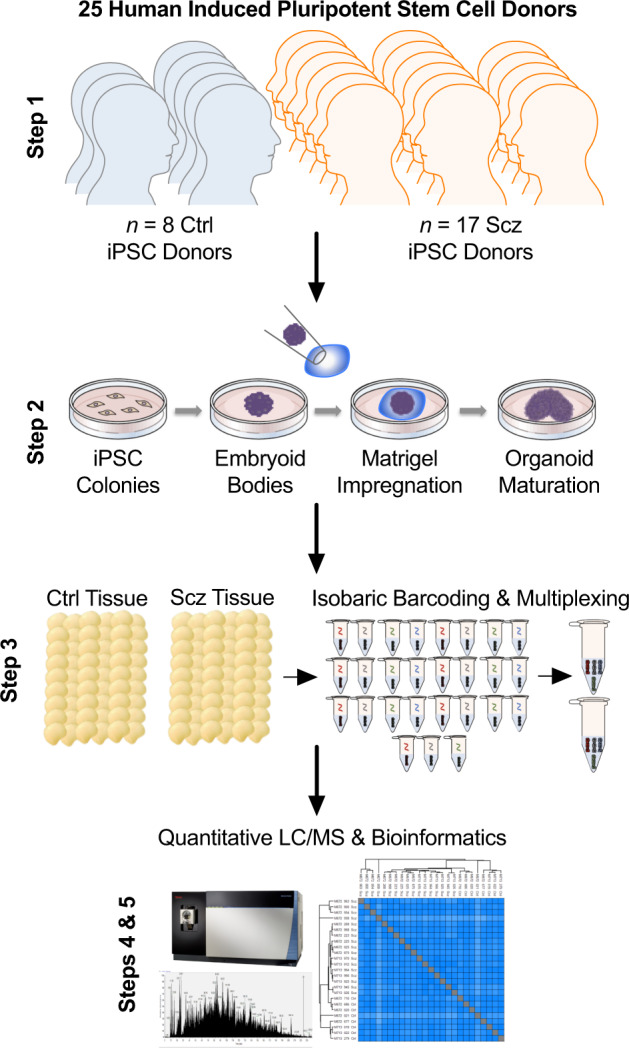
Briefly, 25 distinct human iPSCs were obtained from both healthy Control (Ctrl) donors and Schizophrenia (Scz) patients. Each line represented a biologically unique sample from a specific individual, and lines were predominantly obtained from NIH repositories. Following this, iPSCs were expanded and utilized to generate patient-derived cerebral organoids that mimic the 1st trimester of brain assembly (see Methods, [17, 87] for protocol information, and [1] for our previous application of 3D Scz patient-derived organoids). This process involved dissociating iPSC colonies to generate 3D embryoid body aggregates that could be pushed towards a neural fate via chemically minimalist media cocktails [17, 87]. Following neural induction, organoids were implanted into a matrigel droplet as a scaffold to support tissue expansion and (broadly consistent with our prior study [54]) maturated to a primary endpoint of 35-40 DIV. Following this, samples were individually subjected to protein lysis and tryptic-based enzymatic digestion. For proteomic analysis of cerebral organoids, peptides were isobarically barcoded using TMTpro 16-Plex chemistry that allowed samples to be multiplexed for simultaneous detection of different samples via liquid chomatography–mass spectrometry (LC/MS). This allowed up to 15 samples (+1 pool) to be condensed into a single tube for simultaneous detection via LC/MS analysis, resulting in a total of 27 samples (n = 25 human donor organoids, + n = 2 internal reference pools). Bioinformatics were subsequently conducted in accordance with the parameters described in our Methods as well as two prior manuscripts that have incorporated LC/MS proteomic analysis of human-derived organoid samples [1, 11].
