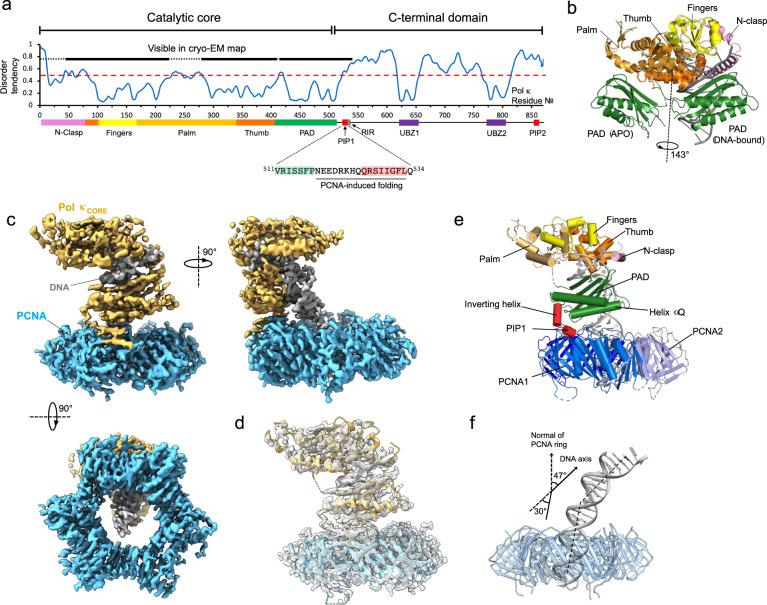
Fig. 1. Cryo-EM structure of the Pol κ−DNA−wt-PCNA complex.
a Disorder prediction against residue number of human Pol κ, and Pol κ domain organization and amino acid sequence of the PCNA-interacting region; PIP PCNA-interacting motif, RIR Rev-1 interacting motif, UBZ Ubiquitin binding zinc-finger. Disorder prediction was performed with PrDOS94. The red dotted line corresponds to a disorder tendency of 0.5. The thick black line corresponds to Pol κ residues observed in the cryo-EM reconstruction. b X-ray structures of apo- (PDB ID 1T94)31 and DNA-bound Pol κ (PDB ID 2OH2)30 overlaid on the core domain. The PAD in the apo structure is rotated 143° relative to the PAD in the DNA-bound structure. Pol κ sub-domains are colored as in panel (a). c Cryo-EM density map of Pol κ complex colored by components (Pol κ in orange, PCNA in skyblue and DNA in gray). d Structural model fitted into the cryo-EM map. e Structure of Pol κ complex colored by domain. f DNA bending in Pol κ holoenzyme model. PCNA is shown as a transparent blue ribbon, DNA as a gray ribbon. Pol κ core and PAD were removed for clarity.