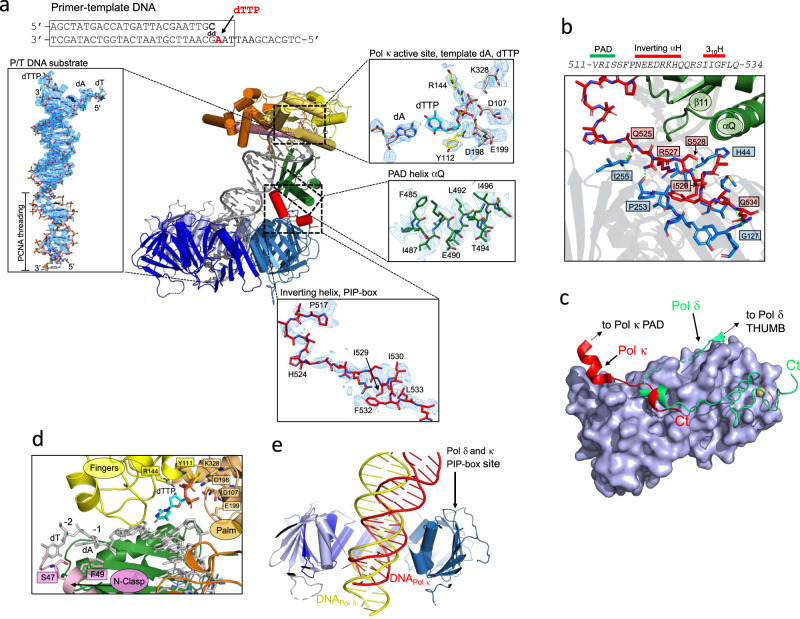
Fig. 2. Details of the cryo-EM structure of the Pol κ−DNA−wt-PCNA complex and comparison with Pol δ processive holoenzyme structure.
a Map region around different elements of the Pol κ−DNA−wt-PCNA complex. The sequence of the DNA P/T substrate is shown. The region of the substrate that was modeled is boxed. b Inter-molecular interactions at the Pol κ−PCNA binding site. Pol κ PIP-box and PCNA-interacting residues are shown as red and blue sticks, respectively. Hydrogen bonds are shown as green dotted lines. Residues forming the canonical PCNA hydrophobic cleft are shown but not labeled. Pol κ PAD and PCNA are shown as ribbons and colored by domain. c Pol κ and Pol δ binding to PCNA. The region of Pol κ and Pol δ interacting with PCNA is shown as red and green ribbons, respectively. Interacting PCNA protomer is shown as a light blue surface. d Model region of Pol κ active site. Pol κ is shown as a ribbon colored by domains, residues interacting with DNA are shown as sticks. DNA is shown as gray sticks. e Side-view of the cryo-EM structures of Pol δ (PDB ID 6TNY)36 and Pol κ holoenzymes aligned on PCNA. PCNA subunits are shown in different shades of blue and the subunit in the foreground is removed for clarity. DNA molecules in Pol δ and κ structures are shown as yellow and red ribbons, respectively.