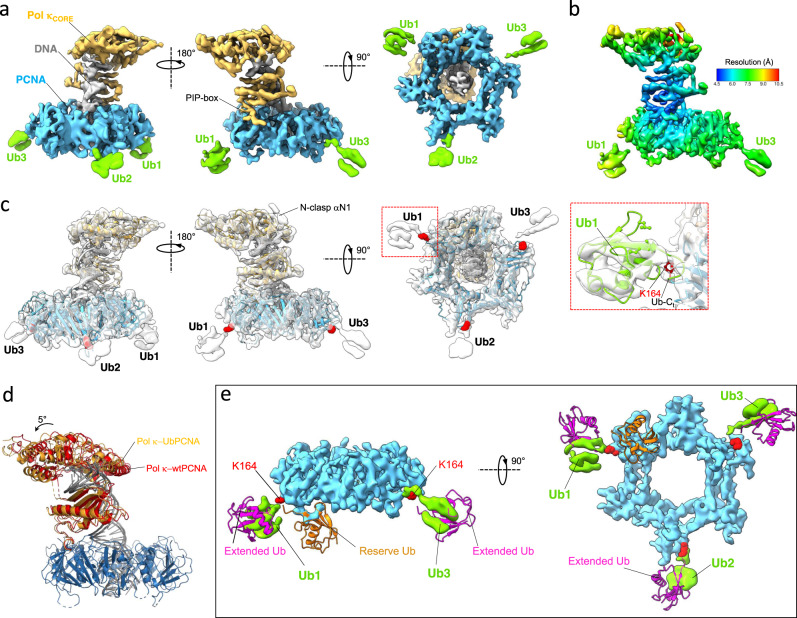
Fig. 4. Cryo-EM structure of the Pol κ−DNA−Ub-PCNA complex.
a Different views of the 6.4 Å reconstruction colored by components (Pol κ in orange, PCNA in skyblue, DNA in gray and ubiquitin in green). b Cryo-EM map color-coded by local resolution. c Structural model fitted into the cryo-EM map. The lysine residues at position 164 are shown as red spheres. The inset shows the rigid-body fitting of ubiquitin structure into the cryo-EM map at Ub1 position. The ubiquitin L8/I44/V70 residues at the hydrophobic patch, and K164 on PCNA are shown as sticks. d Overlay of structural models of Pol κ bound to wt-PCNA or Ub-PCNA aligned on PCNA, highlighting the slight difference in Pol κ core tilt. e Fitting of crystal structures of yeast split Ub-PCNA with ubiquitin in the “reserve” position (PDB ID 3L0W)45, and human Ub-PCNA with ubiquitin in the “extended” position (PDB ID 3TBL)46 into the cryo-EM map of the Pol κ−DNA−Ub-PCNA complex. Cryo-EM densities of PCNA and ubiquitin are colored in skyblue and green, respectively. Map portions of Pol κ and DNA were removed for clarity. The lysine residues at PCNA position 164 are shown as red spheres.