Fig. 2. Repeatable evolution of mutation ntrB A289C is robust across genetic background and nutrient environment (total N = 116).
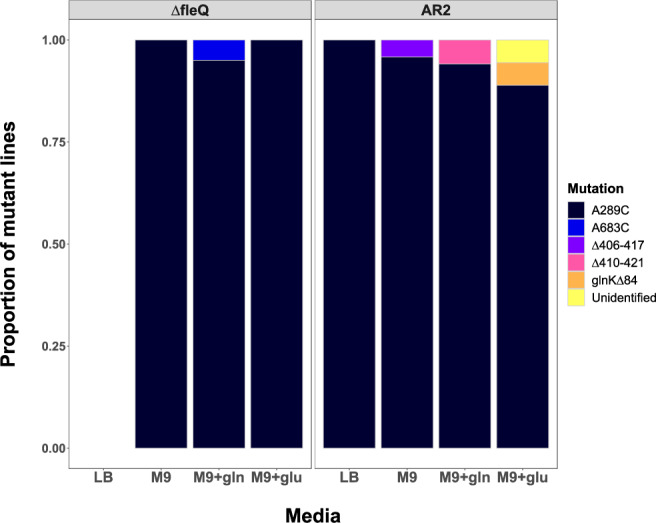
The proportion of each observed mutation is shown on the y axis, and each unique mutational target is highlighted by a colour. Lines were evolved using 4 mM, 8 mM and 16 mM of amino acid supplement glutamate (glu) or glutamine (gln; see Methods). No significant relationship between supplement concentration and evolutionary target was observed (Kruskal–Wallis chi-squared tests, two-sided: AR2 M9 + glu, df = 2, P > 0.2; AR2 M9 + gln, df = 1, P > 0.23; ∆fleQ M9 + gln, df = 1, P > 0.3), as such they are grouped visually and for subsequent statistical analysis. ntrB mutation A289C was robust across both strain backgrounds (SBW25ΔfleQ - shown as ΔfleQ, and AR2) and the four tested nutritional environments, remaining the primary target of mutation in all cases (>88%). As such no significant gene-by-environment interaction was observed (χ2 = 0.875, df = 3, P = 0.83). ΔfleQ lines evolved on LB were able to migrate rapidly through sliding motility alone, masking any potential emergent flagellate blebs (see37). Sample sizes (N) for other categorical variables: ΔfleQ – M9: 25, M9 + gln: 20, M9 + glu: 7; AR2 - LB: 5, M9: 24, M9 + gln: 17, M9 + glu: 18. Source data are provided as a Source Data file.
