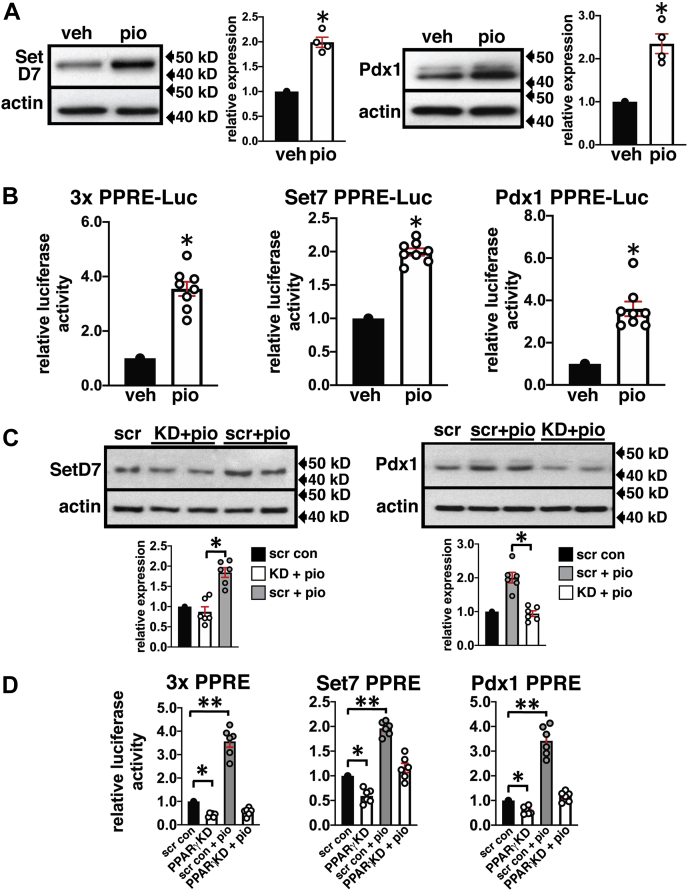
Figure 3.
Pioglitazone-mediated activation of PPARγ upregulates protein expression and promoter reporter activities of SetD7 and Pdx1 that is attenuated in PPARγ KD INS-1 cells.A, representative WB showing increased expression of SetD7 and Pdx1 by the PPARγ agonist-pioglitazone compared with vehicle treated INS-1 cells: INS-1 cells were treated with 10 μM pioglitazone or DMSO for 72 h. Cell lysates were prepared and processed for WB analysis for SetD7 and Pdx1 with β-actin used for protein loading normalization. Band quantitation was performed using NIH-ImageJ, and the data presented is representative of four separate blots of SetD7 and Pdx1. ∗p < 0.01. B, promoter reporter assays for PPARγ, SetD7, and Pdx1: INS-1 cells transfected with 3xPPRE-Luc, SteD7-PPRE-Luc, and Pdx1-PPRE-Luc promoter reporter vectors. The Renilla luciferase reporter plasmid was included in all transfections serving as an internal control. Twenty-four hours post transfection, cells were treated with 10 μM pioglitazone or DMSO for 48 h. Firefly luciferase activity was measured by a luminometer, normalized with Renilla luciferase, and expressed as relative luciferase activity compared with the vehicle-treated experimental group. The data presented are representative of three independent experiments. ∗p < 0.00001. C, representative WB showing induction of SetD7 and Pdx1 protein levels in the scr control INS-1 cells treated with 10 μM pioglitazone for 72 h relative to vehicle (DMSO) control; however, INS-1 cells with PPARγ KD lacked pioglitazone responsiveness to modulate SetD7 and Pdx1 protein levels. The WB quantitation data is representative of n = 6 samples. ∗p < 0.001. D, promoter reporter assays for PPARγ, SetD7, and Pdx1 in PPARγ shRNA INS-1 cells compared with Scr shRNA expressing INS-1 cells treated with vehicle or pioglitazone. ∗p < 0.05, ∗∗p < 0.01.