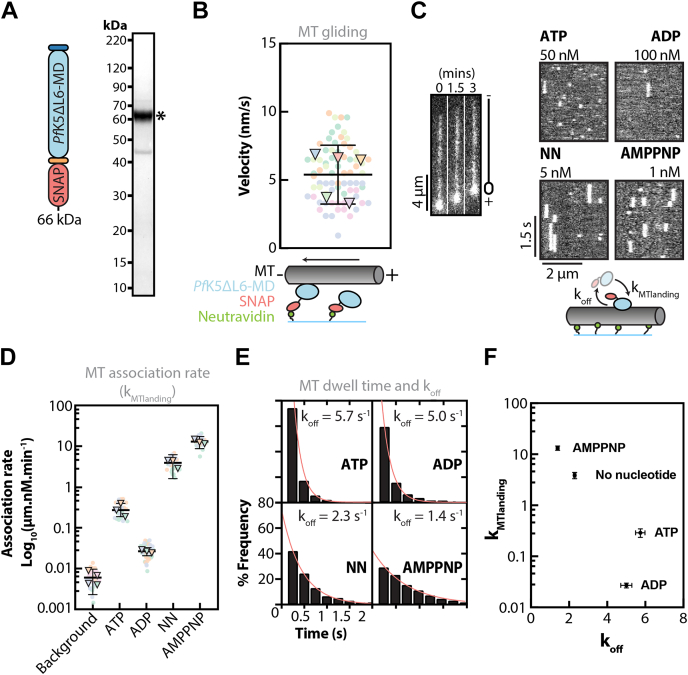
Figure 2.
PfK5ΔL6-MD drives plus-end directed MT gliding and its MT interactions are nucleotide modulated.A, schematic of the PfK5ΔL6-MD-SNAP fusion protein used for TIRFM experiments, and SDS-PAGE of PfK5ΔL6-MD-SNAP after purification. ∗ indicates the band for PfK5ΔL6-MD-SNAP. B, PfK5ΔL6-MD-SNAP driven MT gliding velocity. Technical replicates = 75 (circles), experimental replicates = 5 (triangles), biological replicates = 3. The mean of experimental replicates (5.4 nm/s) and 95% confidence interval are plotted. Grayscale images on the right are snapshots of a single bright plus-end labeled MT over time. C, example kymographs from PfK5ΔL6-MD-SNAP single molecule MT-binding experiments, with each vertical white streak corresponding to a single PfK5ΔL6-MD-SNAP binding event. Concentrations refer to the amount of PfK5ΔL6-MD-SNAP required to see single molecule binding for each nucleotide. D, PfK5ΔL6-MD-SNAP MT association rates in different nucleotide states (NN = no nucleotide). For each nucleotide condition, technical replicates (circles) and experimental replicates (triangles) are plotted, in addition to the mean and 95% confidence interval of experimental replicates. Number of MTs = 34, 22, 46, 28, 29 for the background, ATP, ADP, no nucleotide, and AMPPNP states, respectively. E, frequency distribution of PfK5ΔL6-MD-SNAP MT dwell times, in different nucleotide states. Number of experimental replicates = 3; however, frequency distributions are calculated from pooled experimental data. Number of biological replicates = 2. The fit for one-phase exponential decay models is shown, with corresponding decay constant (koff). Number of events = 1065, 1084, 1799, 1717 for ATP, ADP, no nucleotide, and AMPPNP states, respectively. F, mean MT association (kMTlanding) as a function of MT dissociation (koff), plotted with 95% confidence intervals.