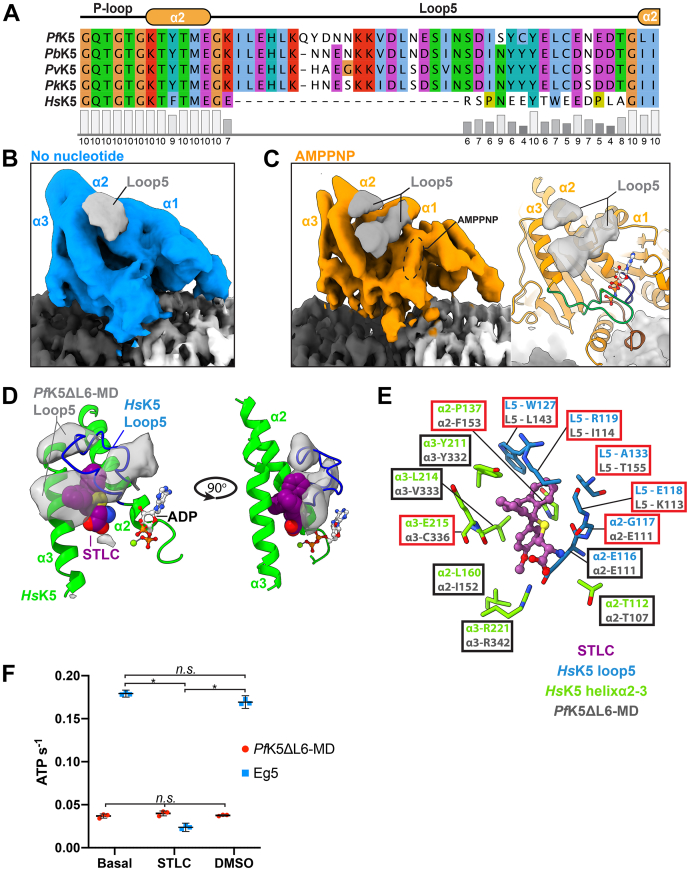
Figure 7.
PfK5ΔL6-MD loop5 alters the environment of the kinesin-5 drug-binding site.A, primary sequence alignment of loop5 from HsK5 (UniProt ID: P52732), PfK5 (O77382) and various other Plasmodium species (Pv = vivax (A0A564ZV10), Pk = knowlesi (A0A1Y3DTC2), Pb = berghei (A0A122I4M3)). Conserved positions are colored according to the ClustalX scheme, and a conservation score as calculated in Jalview is given below. B, Loop5 density (gray) in the no nucleotide state, compared with other PfK5ΔL6-MD density (blue). C, Loop5 density (gray) in the AMPPNP state, compared with other PfK5ΔL6-MD density (left), and the PfK5ΔL6-MD model (right). D, the STLC bound HsK5 crystal structure in lime (PDB ID: 2WOG (89)), which was rigid body fitted into the PfK5ΔL6-MD AMPPNP state map. STLC is colored purple, and HsK5 loop5 is colored blue. PfK5ΔL6-MD loop5 cryo-EM density is depicted in gray. E, conservation of residues partaking in STLC binding between PfK5ΔL6-MD and HsK5, based on primary sequence alignment in (A). Using PDB ID: 2WOG, HsK5 residues contacting STLC were found using Chimera, and are displayed, with loop5 residues colored blue, and residues from helicesα2-3 colored green. Equivalent PfK5 residues are shown in gray with HsK5 residue labels. Nonconserved residues are displayed in red boxes, with conserved ones shown in black boxes. F, ATPase rates of PfK5ΔL6-MD and HsK5 rate in the absence of MTs, with either no treatment, + 20 μM STLC, or DMSO (with the same % v/v as the % v/v of STLC). Statistical relationships were tested using a one-way ANOVA, followed by a post-hoc Tukey's multiple comparison test.