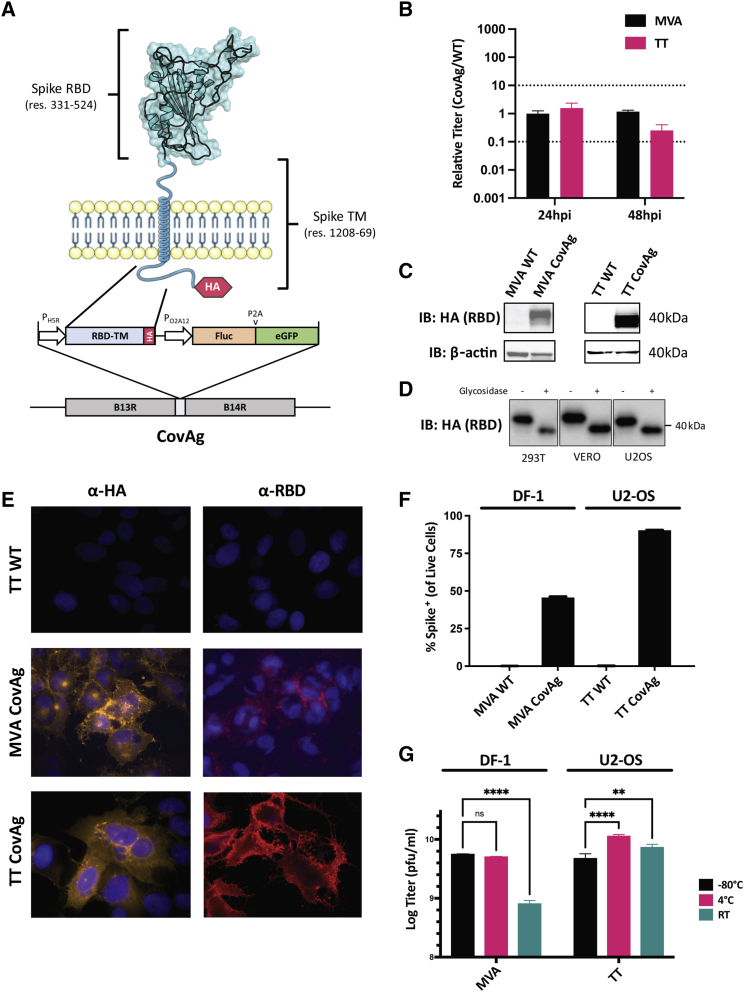
Figure 1.
Design and validation of CovAg vaccine
(A) Schematic of CovAg (spanning residues 331 to 524 and 1,208 to 1,269) antigen49 and its insertion site in the Vaccinia genome. The RBD-TM was designed with a C-terminal HA-tag for detection and is expressed from the Vaccinia H5R promoter (early/late). It was inserted at the B14R locus along with firefly luciferase and GFP for in vivo and in vitro detection, respectively. (B) Comparison of titers from cells infected with MVA CovAg or TT CovAg. Data are shown as CovAg titers relative to WT (n = 3, mean ± SD). Dashed lines reflect 10-fold increase or decrease in points. (C) Immunoblot of CovAg expressed from U2OS cells infected MVA and TT as probed with HA antibody. (D) Immunoblot of CovAg from infected U2OS cells after undergoing treatment with glycosidase to illustrate RBD glycosylation (uncropped western can be found in Figure S2). (E) Immunofluorescence of MVA/TT CovAg constructs with α-HA or α-RBD. For HA antibody samples, cells were permeabilized with 0.2% Triton X-100, whereas RBD samples were left unpermeabilized. (F) Quantification of RBD expressed on the surface of live cells infected with MVA- or TT- CovAg, by flow cytometry (n = 3, mean ± SD). (G) Temperature stability of MVA and TT backbones as probed by plaque assay after storage at −80°C, 4°C, or room temperature (RT) for 7 days (n = 3, log-transformed titer means ± SD; two-way ANOVA with Sidak's correction for multiple comparisons; alpha threshold = 0.05).