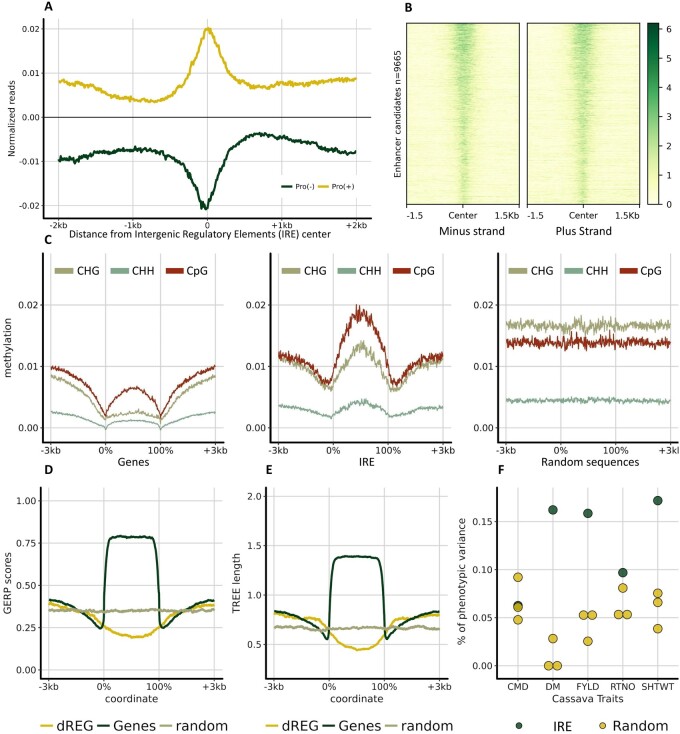
Figure 2.
IREs in cassava have a particular methylation pattern, are evolutionary less conserved and explain more phenotypic variance than expected for several agronomic traits. (A) Pro-seq reads mapping around cassava IREs. Reads were sorted by strand and the normalized reads were plotted around the center of each candidate. (B) Heatmap representation of reads mapping to the enhancer candidate regions. The regions are sorted based on dREG scores. (C) Cassava methylation patterns for the three methylation contexts (CG, CHH, and CHG) were plotted around the genic regions, IREs, and a set of random sequences. The random set has the same number and length distribution as the IREs. Genomic regions were scaled (0–100%) for visualization. (D) GERP scores and corresponding tree lengths (E) were also plotted around the IREs (dREG), Genes, and random set of regions. (F) Genomic Partitioning of complex agronomic traits (DM: Dry matter content; FYLD: Fresh yield; RTNO: Root number; SHTWT: Shoot weight) and a disease trait (CMD: Severity of Cassava Mosaic Disease). Relationship matrices were calculated using SNP markers within the enhancer candidate regions using the LDAK5 model and variance components were estimated using EMMREML.