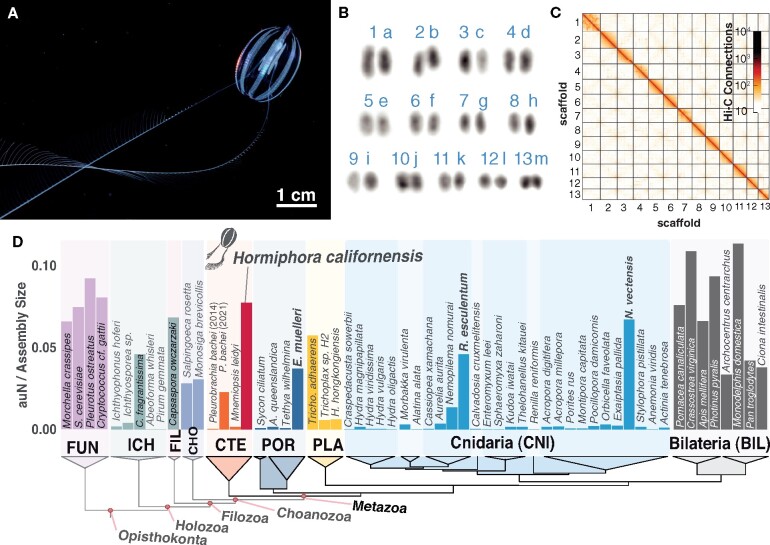
Figure 1.
Karyotype and genome assembly quality. (A) H. californensis with its tentacles extended during feeding. (B) One karyotype image (rearranged and color-inverted) of a H. californensis chromosome preparation that contained 13 chromosome pairs. (C) The H. californensis Hi-C map, showing thirteen chromosome-scale scaffolds. (D) Genome contiguity normalized by genome size [auN/Assembly Size, per (Li 2020)] from all available nonbilaterian holozoan genomes and select fungal and bilaterian genomes, with tree topology based on (Whelan et al. 2017). Bolded species names are nonbilaterians with chromosome-scale genome assemblies. BIL, bilaterians; CHO, choanoflagellates; CNI, cnidarians; CTE, ctenophores; FIL, Filasteria; FUN, Fungi; ICH, Ichthyosporea; PLA, placozoans; POR, Porifera.