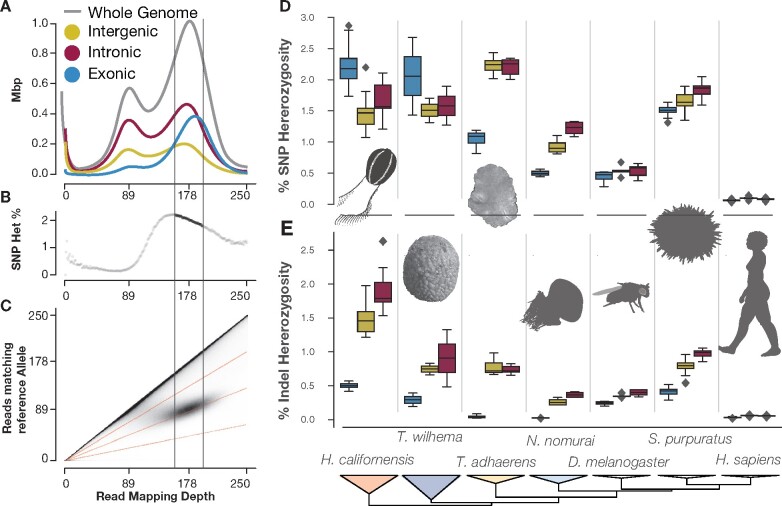
Figure 3.
The Hormiphora genome is highly heterozygous and contains many indels in noncoding regions. (A) The histogram of the number of bases in a genome assembly (y-axis) with a specific mapping depth (x-axis) using Illumina WGS reads. The mode of the mapping depth of the Illumina WGS reads was 178x, so bases with close to 178x mapping coverage have reads from both haplotypes. Therefore, bases with a mapping coverage around 89x have reads mapped from a single haplotype. (B) The whole-genome heterozygosity calculated at each read mapping depth. The heterozygosity value for each mapping depth (x-axis) is the number of sites where only one half of the mapped reads match the reference allele, divided by the total number of sites at that depth. (C) The 2D histogram showing that bi-allelic sites are centered around 178x read mapping depth. Also, the 2D histogram is one way to visualize the data input for panel (B). (D–E) Box-and-whiskers plots show the distribution of SNP (D) or indel (E) heterozygosity in each chromosome-size scaffold. In H. californensis, the intergenic and intronic regions have a reduced SNP heterozygosity, but an elevated rate of indels. This pattern differs from the correlation between SNP and indel heterozygosity found in species with lower overall heterozygosity.