Figure 4.
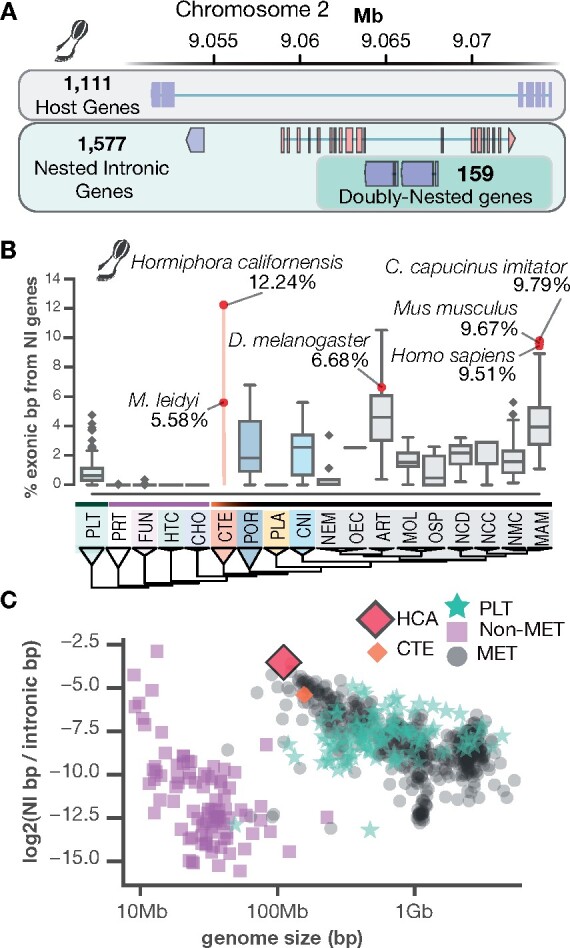
An analysis of NI genes in 1058 eukaryote genomes. (A) NI genes are genes that are contained within the introns of other genes. In this example from the H. californensis genome, four genes are nested within a single host gene. Two of the genes are doubly nested—they are within the intron of another NI gene. Blue genes are coded on the antisense strand relative to the reference sequence and red genes are coded on the sense strand. (B) The percent of all exonic bp that are in NI genes. Protists and nonmetazoans have a negligible proportion compared to animals. Abbreviations: ART, panarthropoda; CHO, choanoflagellates; CNI, cnidarians; CTE, ctenophores; FUN, fungi; HCA, H. californensis; HTC, holozoa through choanoflagellates; MAM, mammals; MET, Metazoa; MOL, molluscs; NCD, nonchordate deuterostomes; NCC, noncraniata chordates; NEM, nematodes; NMC, nonmammalian chordates; OEC, other ecdysozoa; OSP, other spiralians; PLA, placozoans; PLT, plants; POR, Porifera; PRT, other protists. (C) As animal and protist genomes become smaller, a higher proportion of the exonic bases are in NI genes.
