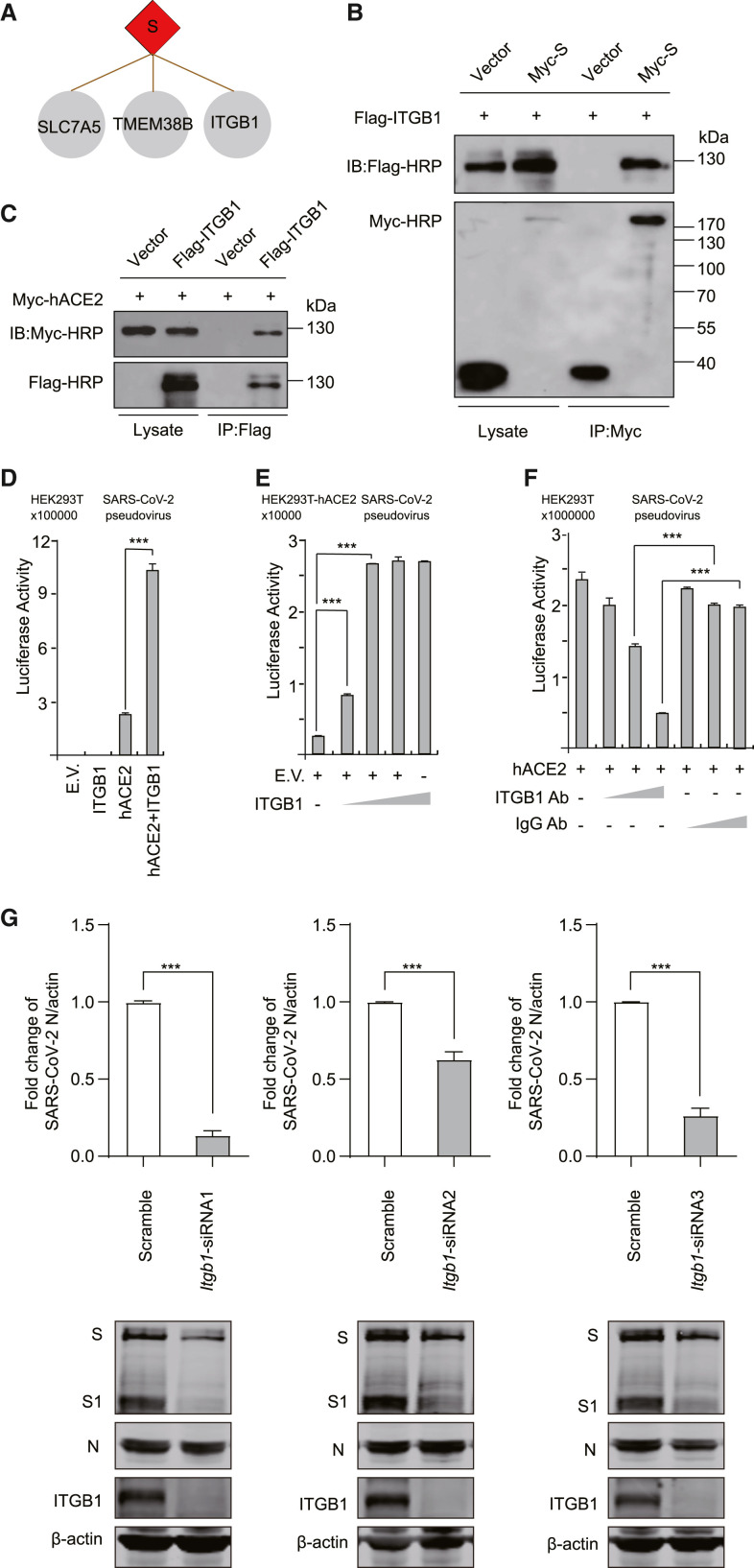
Figure 4.
ITGB1 mediates the entry of SARS-CoV-2
(A) The binding partners of the S protein.
(B) Validation of the interactions of ITGB1 and S by co-immunoprecipitation assay. HEK293T cells were co-transfected with ITGB1 and S, immunoprecipited by Myc antibody, and detected by Myc or Flag antibodies.
(C) Validation of the interactions of ITGB1 and hACE2 by co-immunoprecipitation assay. HEK293T cells were co-transfected with ITGB1 and hACE2, immunoprecipited by Flag antibody, and detected by Myc or Flag antibodies.
(D) HEK293T cells were transfected with ITGB1 and hACE2. Then, the cells were infected with SARS-CoV-2 pseudovirus.
(E) HEK293T-hACE2 cells were transfected with gradually increased ITGB1. Then, the cells were infected with SARS-CoV-2 pseudovirus.
(F) HEK293T cells were transfected with hACE2. Then, the cells were infected with SARS-CoV-2 pseudovirus and treated with anti-ITGB1 antibody or scramble antibody.
(G) HeLa-hACE2 cells were transfected with scramble or ITGB1-specific siRNA oligos as indicated. The cells were infected with SARS-CoV-2. Total RNA was analyzed using real-time quantitative PCR (RT-qPCR) to determine the expression levels of the N protein. The cell lysates were analyzed using western blotting as indicated. β-Actin was used as a loading control.
∗p < 0.05, ∗∗∗p < 0.001 (two-tailed Student's t test), means + SD, n = 3. Data are representative of two (B and C) or three (D–G) independent experiments. E.V., empty vector.