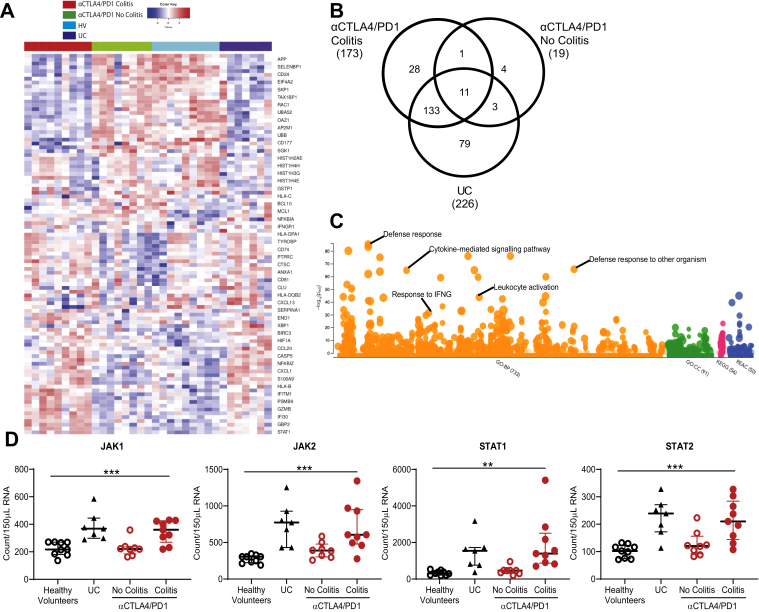
Figure 3.
Targeted gene panel analysis of ICI-colitis includes unique and common features compared with UC and high expression of the IFNG signaling pathway. A 780-gene Nanostring analysis of colonic biopsy RNA from HVs (n = 8), patients with active UC (n = 5), DCC (n = 9), and DCNC (n = 8). (A) Heatmap of top 50 differentially expressed genes. (B) Number of genes up-regulated ≥2-fold compared with HVs demonstrates 173 of up-regulated genes in DCC, only 12 of 173 are also up-regulated in the DCNC group (limited on-treatment effect); 144 of 173 genes are common between DCC and UC, 28 of 173 genes are unique to DCC. (C) Manhattan plot indicating significantly up-regulated pathways, including response to IFNG. The complete list is provided in Supplementary Table 4. (D) RNA expression of canonical markers of IFNG signaling JAK1, JAK2, STAT1, and STAT2 are higher in DCC and UC groups compared with healthy controls and DCNC groups. ∗∗P < .01 and ∗∗∗P < .001 by 1-way analysis of variance.