Figure 2. RBD-scNP immunization elicits higher titers of nAbs against more transmissible or neutralization-resistant SARS-CoV-2 variants than stabilized spike mRNA-LNP vaccination.
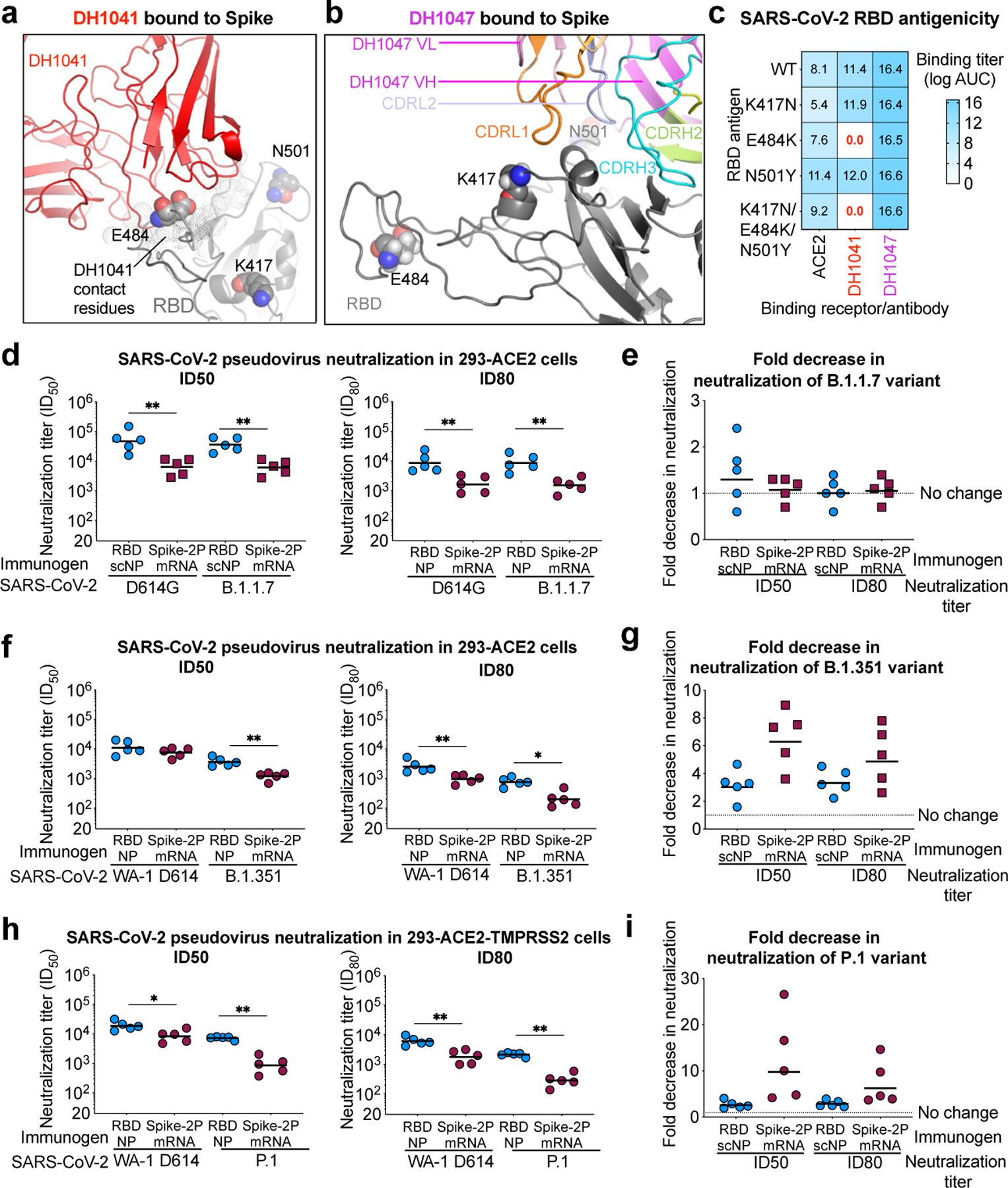
a,b The location of K417, E484, and N501 (spheres) present in the B.1.351 variant are shown in the cryo-EM structures of RBD nAbs a DH1041 (red) and b DH1047 (magenta) bound to the RBD (gray) of S trimers (PDB: 7LAA and 7LD1)15.
c ACE2 receptor, DH1041, and DH1047 ELISA binding titer as log AUC for wildtype and mutant SARS-CoV-2 Spike RBD monomers.
d Serum neutralization ID50 (left) and ID80 titers for SARS-CoV-2 D614G and SARS-CoV-2 B.1.1.7 pseudoviruses from immunized macaques. Symbols represent individual macaques and horizontal bars are group means (**P = 0.0079, Two-tailed Exact Wilcoxon test, n = 5 macaques).
e Fold decrease in neutralization potency between neutralization of SARS-CoV-2 D614G and SARS-CoV-2 B.1.1.7 pseudoviruses. Fold change is shown for RBD-scNP-immunized and mRNA-LNP-immunized macaques based ID50 (left) and ID80 (right) titers. Horizontal bars are the group mean.
f Vaccinated macaque serum neutralization ID50 (left) and ID80 titers (right) against WA-1 and B.1.351 pseudovirus. Symbols and horizontal bars are shown the same as in d (*P = 0.0159 and **P = 0.0079, Two-tailed Exact Wilcoxon test, n = 5 macaques).
g Fold decrease in neutralization potency between neutralization of SARS-CoV-2 WA-1 and B.1.351 pseudoviruses. Fold change is shown the same as in e .
h Vaccine-induced neutralization of SARS-CoV-2 WA-1 and P.1 pseudovirus infection of ACE2 and TMPRSS2-expressing 293 cells. Symbols and horizontal bars are shown the same as in d (*P = 0.0159 and **P = 0.0079, Two-tailed Exact Wilcoxon test, n = 5 macaques).
i Fold decrease in neutralization potency as shown in e between neutralization of SARS-CoV-2 WA-1 and P.1 pseudoviruses.
