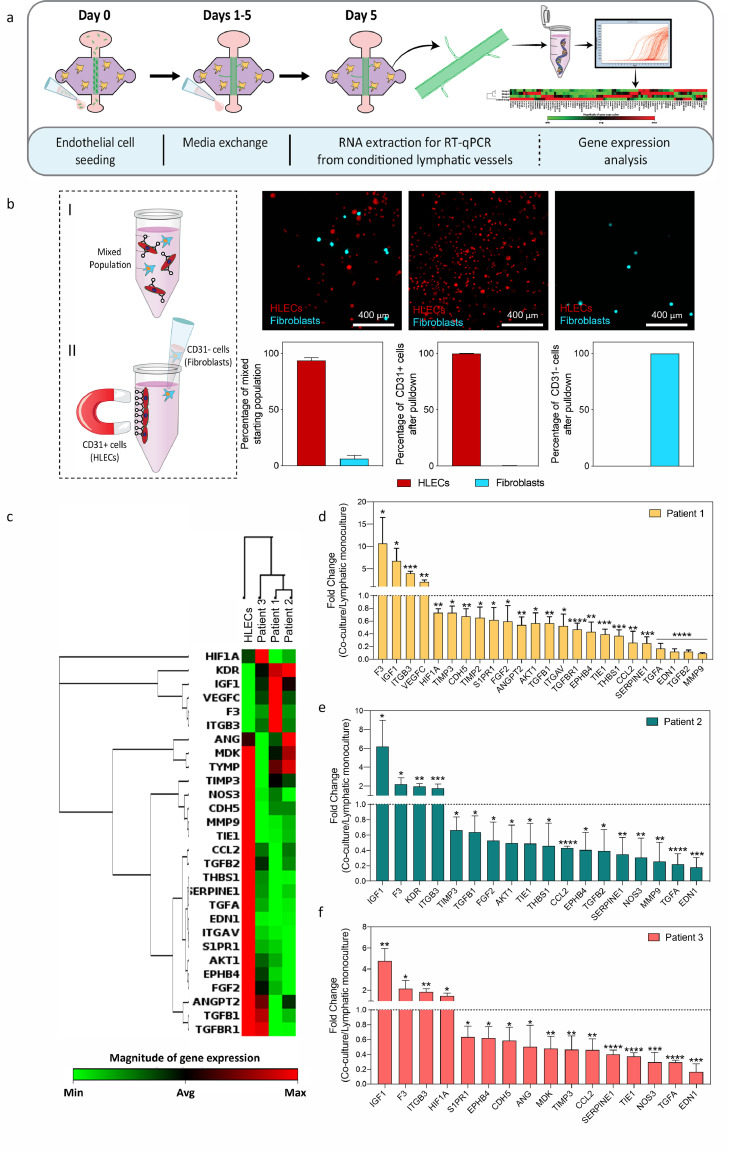
Figure 4.
Results of qPCR analysis from the conditioned lymphatic vessels. a) Schematic indicating the culture procedure and the procedure to obtain mRNA from the conditioned lymphatic vessels at day 5 for gene expression analysis. b) Schematic of HLECs cell separation. HLECs cells attach to CD31 magnetic beads, supernatant is discarded, thus isolating HLECs cells. Confocal images represent captured HLECs cells (in red) and discarded fibroblasts (in blue). The graph highlights the efficiency of HLECs cell isolation. N = 3 independent biological replicates. c) Unsupervised clustergrams were produced using only those genes showing significant changes. N = 3 independent biological experiments per patient with 4 pooled lumens each (technical replicates used to obtain enough mRNA for analysis). d-f) Significant changes in gene expression for each patient were plotted for patients 1, 2, and 3 to reflect the direction of the change. Bars represent average ± S.D. p ≤ 0·05, **p ≤ 0·01, ***p ≤ 0·001, ****p ≤ 0·0001 via one-way ANOVA + Dunnett post-hoc test.