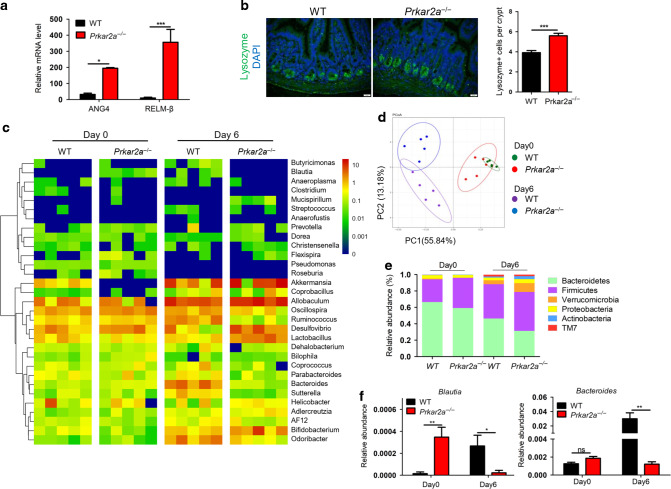
Fig. 9. Ablation of Prkar2a alters gut bacterial composition.
a RT-qRCR analysis of the transcription level of ANG4 and RELMβ in colon tissues collected from Prkar2a−/− (n = 3) and WT (n = 3) mice. Data are representative of three independent experiments conducted in duplicate. b Lysozyme immunostaining shows the increased Paneth cells in Prkar2a−/− mice (n = 3) and WT mice (n = 3). Quantification of Lysozyme-positive cells per crypt. Five images per mouse were quantified. Data are representative of two independent experiments. c Heatmap depicting of relative abundance of microbiota at the family level in colon contents collected from Prkar2a−/− and WT mice at day 0 or day 6 of DSS-induced colitis (n = 5 per group). d Principal coordinate analysis (PCoA) of weighted UniFrac distances based on 16S rDNA analysis of microbiota of colon contents from Prkar2a−/− and WT mice pre- and post-DSS challenge (n = 5 per group). Symbols represent data from individual mice. e Relative abundances of microbial commensal diversity were analyzed at the phylum level by 16S rRNA gene sequencing. f Relative abundances of Blautia genera and Bacteroides genera in Prkar2a−/− mice versus WT mice before and after DSS challenge. Data are presented as mean ± SEM. Student’s t test was used to do the analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ns not significant.