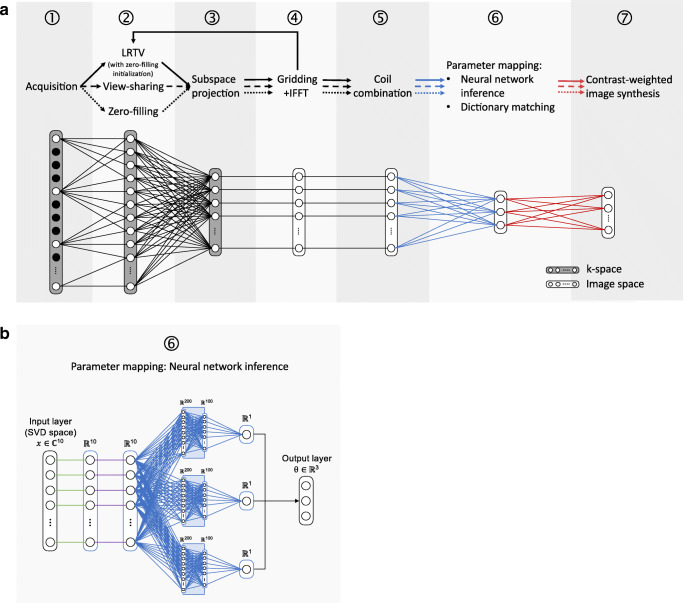
Fig. 1.
3D QTI data processing. a Reconstruction and processing. After acquisition (➀), raw k-space data is processed via naïve zero-filling (dotted line), k-space weighted view-sharing (dashed line), or a compressed sensing LRTV technique (solid line). All methods in ➁ are followed by dimensionality reduction via SVD subspace projection in the time domain (➂), gridding onto a Cartesian grid followed by a 3D IFFT (➃), and coil sensitivity estimation and combination (➄). The reconstructed image series are then fed into a neural network or are matched to a precomputed dictionary to output parametric maps of T1, T2 and PD (➅). We then synthesize clinical image contrasts using the parametric maps (➆). b Neural network architecture for parameter inference. The model receives the complex, voxel-wise signal in SVD subspace x and infers the underlying tissue parameter vector θ with T1, T2, and a PD-related scaling factor. The input signal x is phase-aligned (green lines) to transfer the complex into real-valued signal, followed by a normalization layer (purple lines). The model then divides into separate pathways, each with three ReLU-activated hidden layers and 200, 100, and one node, to eventually yield the concatenated parametric output vector θ