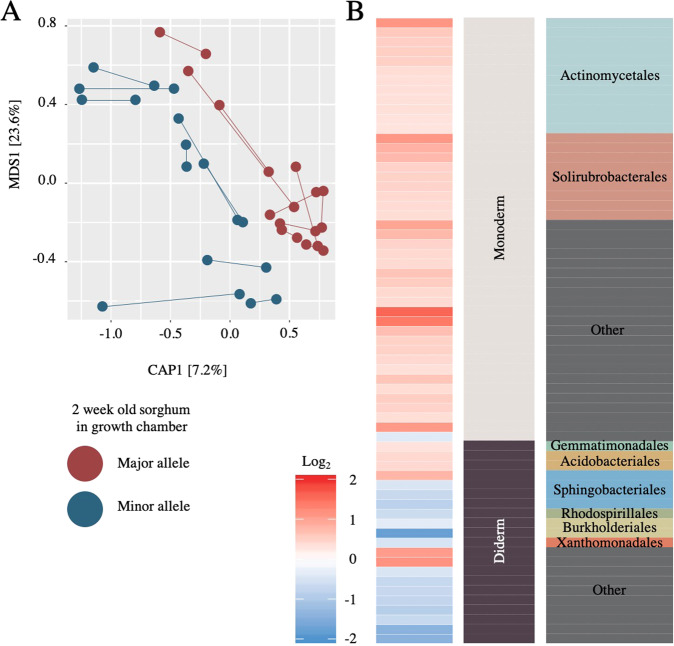
Fig. 5. Sorghum genetic information can be used to predict rhizosphere microbiome composition under different growth conditions.
A Canonical analysis of principal coordinates of the rhizosphere microbiome for nine major allele genotypes (red) and nine minor allele genotypes (blue) grown in a growth chamber for 2 weeks. Two replicates per genotype were used and are connected by lines. B For each indicator OTU, the log2 fold change in abundance between the sorghum major (red) or minor (blue) allele groups was determined. OTUs were grouped based on the predicted presence of one or two membranes (monoderm or diderm), within each bacterial order, and colored as in Figs. 2 and 4.