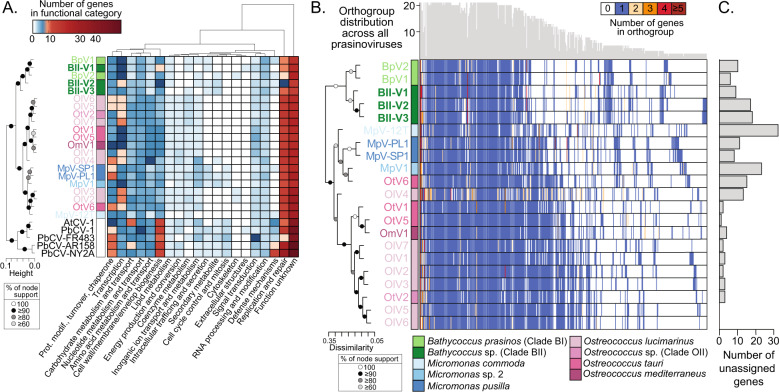
Fig. 2. Distribution of functions and orthologous protein families across genome-sequenced prasinoviruses.
A Functional category distributions across 21 genome-sequenced prasinoviruses and chloroviruses based on EggNOG categorisation. Viruses are clustered by similarity in their distribution of the functional categories on the y-axis and the frequency of each category across the viral genomes determines clustering along the x-axis ordering. Genes with homology to proteins in the EggNOG database but could not be assigned a function are in the “function unknown” category. B Orthogroups presence/absence patterns ordered along the x-axis by ranking according to the total number of genes in the orthogroup. For inclusion, the orthogroup was required to include protein sequences from at least two different viral genomes. Viruses are ordered along the vertical by their presence/absence pattern reconstructed by hierarchical clustering (topology on the left). Top histogram: frequency of each orthogroup in sequenced prasinoviruses. C Genes in each virus (number) not assigned to any orthogroup, with viruses in the same vertical order as B.