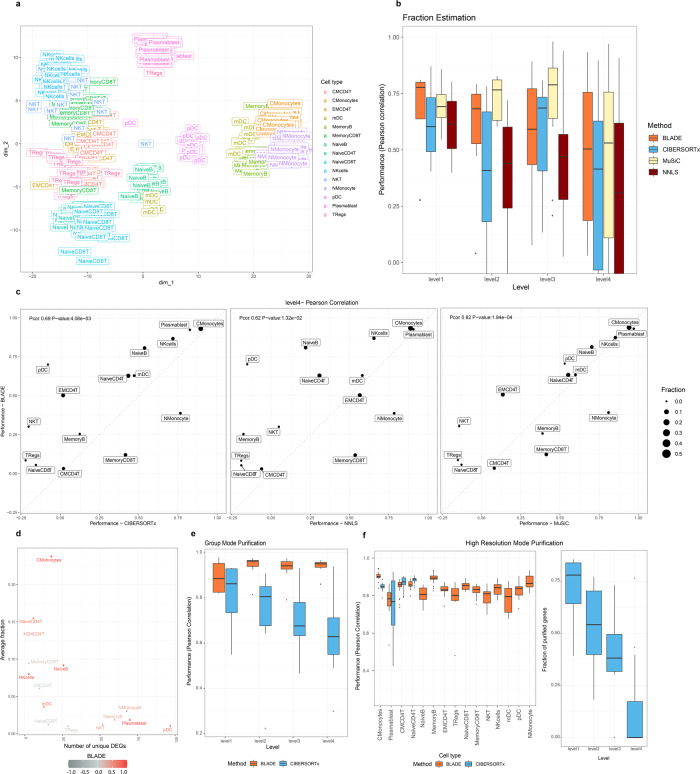
Fig. 5. Performance evaluation of BLADE using simulated PBMC bulk RNA-seq data.
a A t-SNE plot represents the similarities in Pearson correlation coefficients among gene expression profiles of 15 cell types* (denoted by label) in 20 simulated bulk PBMC data. b Performances (Pearson correlation coefficient; y-axis) of BLADE (orange), CIBERSORTx (blue), NNLS (dark red), and MuSiC (light yellow) in predicting cellular fractions of the 20 simulated PBMC bulk RNA-seq data with diverse levels (n = 4, 7, 12, and 15 cell types, respectively, in levels 1–4; x-axis). The standard boxplot notation was used (lower/upper hinges —first/third quartiles; whiskers extend from the hinges to the largest/lowest values no further than 1.5 * inter-quartile ranges). c Comparison of performance in estimating the cellular fractions per cell type of BLADE (y-axis) with CIBERSORTx, NNLS, and MuSiC (x-axis) at level 4. The fraction of each cell type is indicated by the size of the point. Pearson correlation coefficient and two-tailed test P-values are indicated at the top left in each panel. d Performance of BLADE (indicated by color) and its association to the number of unique DEGs per cell type (x-axis) and the respective fraction in the simulated data (y-axis). e Performance in Pearson correlation coefficient of BLADE (orange), CIBERSORTx (blue) for group mode purification of four levels of PBMC simulation data (n = 4, 7, 12, and 15 cell types, respectively, in levels 1–4; x-axis). The standard boxplot notation was used. f Performance (Pearson correlation coefficient; y-axis) of BLADE (orange) and CIBERSORTx (blue) in estimating gene expression profiles per cell type (x-axis) and per sample in level 4 (n = 20 samples per cell type; left). Fraction of genes in silico purified in high-resolution mode by CIBERSORTx at all levels of PBMC simulation data (n = 20 samples with 4, 7, 12, and 15 cell types, respectively, in levels 1–4; x-axis; right). The standard boxplot notation was used. *(CMCD4T: central memory CD4+ T cell; CMonocytes: classical monocytes; EMCD4T: effector memory CD4+ T cell; mDC: myeloid dendritic cell; MemoryB: memory B cell; MemoryCD8T: memory CD8+ T cell; NaiveB: naive B cell; NaiveCD4T: naive CD4+ T cell; NaiveCD8T: naive CD8+ T cell; NKcells: natural killer cell; NKT: natural killer T cell; Nmonocyte: non-classical monocyte; pDC: plasmacytoid dendritic cell; TRegs: regulatory T cell).