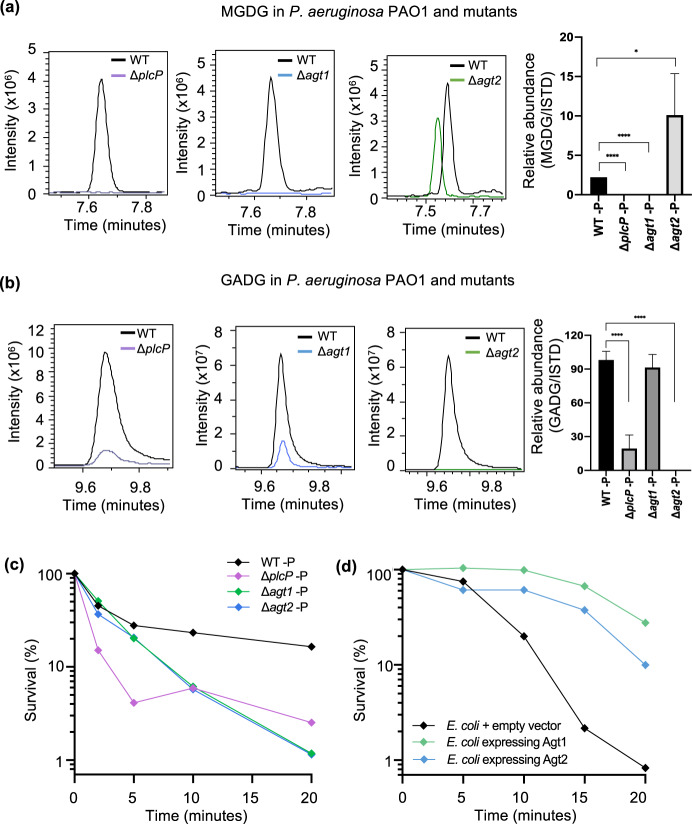
Fig. 5. Glycolipid formation in P. aeruginosa and mutants under Pi stress, showing a protective role of glycolipids to polymyxin B.
Relative abundance of the glycolipid MGDG (a) and GADG (b) in P. aeruginosa mutants plcP (purple trace), agt1 (blue trace) and agt2 (green, trace) compared to the wild type (WT). Cells were cultivated under low Pi conditions (50 µM) and a representative extracted ion chromatogram of MGDG/GADG is shown between the WT, (black trace) and each mutant. The right most panel shows the abundance of MGDG or GADG calculated relative to an internal lipid standard d17:1/12:0 sphingosylphosphoethanolamine (Sigma-Aldrich) in the wild-type and mutant strains of P. aeruginosa. Values are calculated from three biological replicates and the error bars denote standard deviation. MGDG, monoglucosyldiacylglycerol, GADG, glucuronic acid-diacylglycerol. c Survival of glycolipid remodelling mutants ΔplcP (purple), Δagt1 (green) and Δagt2 (blue) when challenged with 4 µg mL−1 polymyxin B compared to WT under Pi stress (black). All experiments were conducted under Pi stress conditions and the results are the average of three biological replicates; error bars denote standard deviation. d Survival of glycolipid producing Escherichia coli when challenged with 20 µg mL−1 polymyxin B. All experiments were conducted in three replicates and error bars denote standard deviation. Black, E. coli containing the empty vector pET28a; green, E. coli containing plasmid pET28a-Agt1; blue, E. coli containing plasmid pET28a-Agt2.