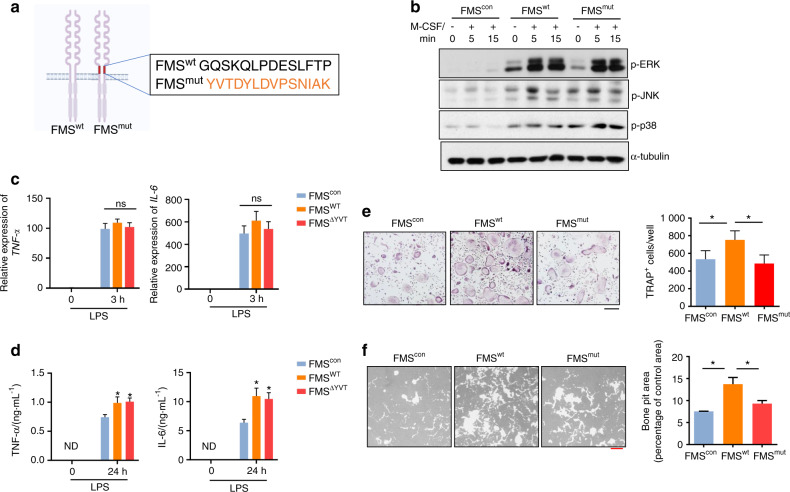
Fig. 3.
c-FMS proteolysis positively regulates osteoclastogenesis. a Schematic showing mutations in the TACE cleavage sites of c-FMS. TACE cleavage sites of c-FMS10 were replaced by the addition of 14 amino acids from insulin receptor sequences (FMSmut). b 293 T cells did not express c-FMS and were transduced with lentiviral particles encoding control, FMSwt or FMSmut. The cells were then stimulated with M-CSF for the indicated times. Protein expression of phospho-ERK, phospho-JNK, phospho-p38, and α-tubulin was determined by immunoblot analysis. c–f BMDMs from c-FMSf/+ Mx1-Cre mice were transduced with lentivirus encoding control, wild-type FMS (FMSwt), or the TACE-uncleavable mutant FMS (FMSmut). The transduced BMDMs were stimulated with LPS (10 ng·mL−1) for 3 h (c) and 24 h (d). c The mRNA expression of TNFα and IL6 was measured by q-PCR. d TNFα and IL6 protein levels in the culture media were measured by Luminex multiplex cytokine assays. e Osteoclastogenesis assay. The left panel shows representative images of TRAP-stained cells. The right panel shows the percentage of TRAP-positive multinuclear cells (MNCs: more than three nuclei) per control (n = 6). Black scale bar: 100 μm. f Resorption pit assay. Bone resorption activity analysis of FMScon, FMSwt, or FMSmut cells. The left panel shows representative images, and the right panel shows the percentage of the resorbed pit area per total area. Red scale bar: 200 μm. All data are shown as the mean ± SEM. ns not significant, ND not detected. *P < 0.05 by one-way ANOVA with a post hoc Tukey test (c–f). The data represent at least three experiments (b–d, f).