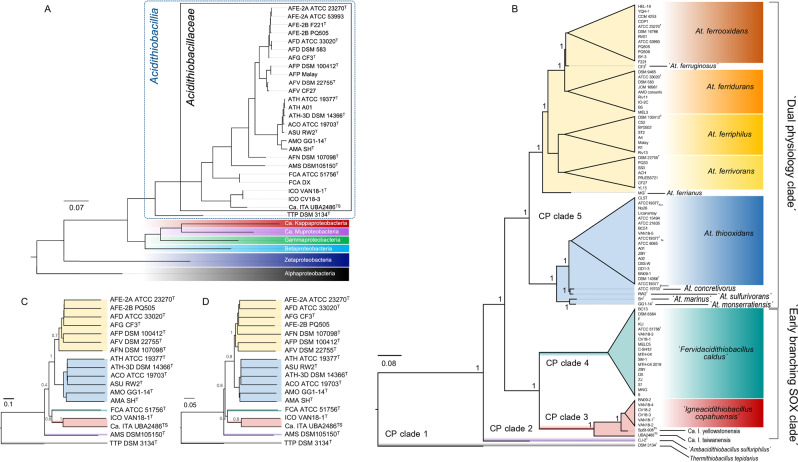
Fig. 2. Acidithiobacillia class consensus phylogenetic trees built using the concatenated alignment of shared genes.
A The MCC Bayesian tree was obtained using MrBayes with a concatenated alignment of 16 RPs (encompassing 2416 aa) that are present in all lineages and outgroups in the analysis. The equivalent tree obtained with all the 16 RPs of the Acidithiobacillia class is presented in Fig. S1d. Alpha- and Zetaproteobacteria form basal clades, consistent with the early branching position of these classes in the evolution of Proteobacteria [93]. B The ML tree was obtained with the PhyML package using 107 conserved single-copy proteins common to 88 genomes of the Acidithiobacillia class. Genomes of seven strains (BY-02, DMC, JYC, S10, DLC-5, GGI-221, and 21-59-9) were excluded from the phylogenomic analysis due to assembly quality issues. The alignment encompassed 24,335 aa and 10,269 parsimony informative sites. C, D Single-gene trees for RpsC (S3) and RplD (L4) constructed using NJ (10000 replicates) are shown in C and D, respectively. RPs and CPs were recovered from the genome assembly versions listed in Table S1b and detailed in Tables S5 and S6. CPs were accrued with an iterative process of filtering for genome-wise occurrence, copy number variations per genome, gene integrity, and length. Sequences aligned in this figure were reduced to simplify tree representation without altering the topology found with comprehensive alignments (Fig. S1). Branches of interest are highlighted in the tree and labeled accordingly. Posterior node support is shown as fractions. Lineages are color coded according to their taxonomic affiliation as elsewhere in the manuscript.