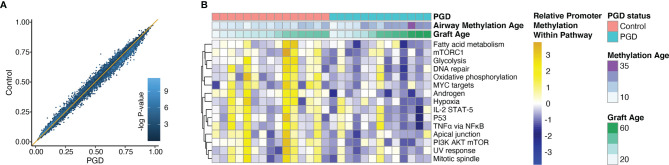
Figure 4.
Promoters within specific gene sets were hypomethylated relative to non-PGD subjects. Average methylation values were determined for CpG sites in gene promoter regions. Average values for PGD subjects (x-axis) are plotted against average values non-PGD control subjects (y-axis) for each CpG site. There were no differentially methylated promoters after adjustment for multiple comparisons. Each dot color corresponds to the -log of the unadjusted p-value for each site (A). Promoter methylation within the MSigDB Hallmark gene sets were compared for between PGD and control subjects. The top 15 most differentially methylated pathways are shown, all with a false discovery-adjusted P-value of <0.01 by Kolmogorov–Smirnov test (B).