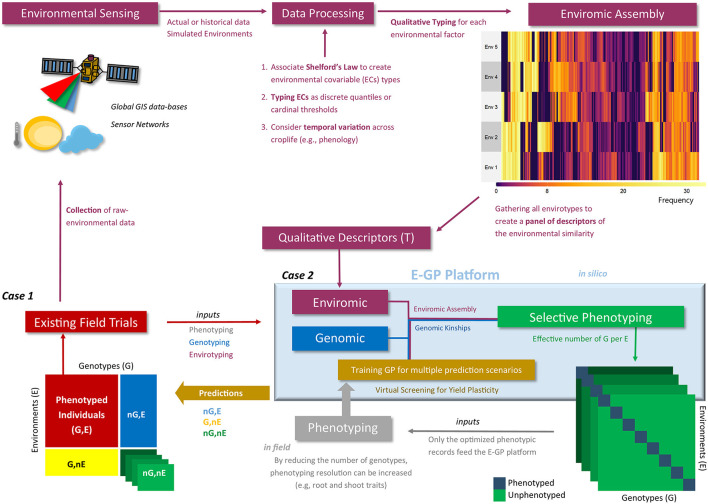
Figure 2.
Workflow of the enviromic-aided genomic prediction (E-GP) considering the two study cases (Case 1 and Case 2) of this study. Phenotypic records from existing field trials (red box) are based on observed genotypes (G) in tested growing environments (E). Currently, these data are being used for training prediction models considering untested genotypes at the same conditions (nG, E), especially when we have some type of structure of genetic relationships, such as genomic data (blue colors). In addition, novel growing conditions can be predicted (G, nE and nG, nE) using enviromic sources (wine colors), Case 1. First, raw environmental data are collected from trials involving equipment installed in situ (e.g., micro-weather stations) or remote sensing techniques. Then the raw data are processed and translated into an enviromic source that carries some ecophysiology process or statistical distribution of the raw data across time and space. The enviromic assembly is then finalized, in which its product is a matrix of envirotype markers by environments. Taking the T matrix as an example (qualitative descriptors based on typologies), a predictive breeding tool merging genomic, enviromics, and phenotypic data can be trained and deliver predictions for several scenarios of G × E. However, there is a second way to create an E-GP platform, the hereafter Case 2, in which the previously collected genomic and enviromic sources for a given TPE are used to develop in silico realizations of the expected G × E for a certain experimental network. Then, optimization algorithms are used to design a selective phenotyping strategy (green box) in which only the most representative genotype-environment combinations are phenotyped and considered for training the E-GP models (gray box). Finally, diverse G × E can also be predicted.