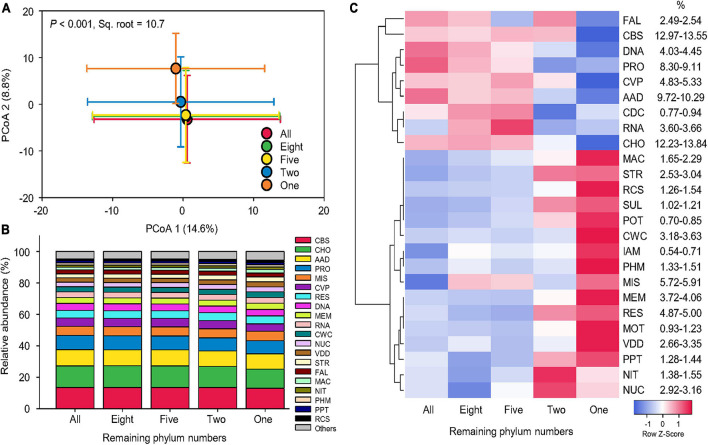
FIGURE 4.
Stable functional structure. (A) Principal coordinate analysis (PCoA) showing the beta-diversity of functional profiles at the genus level affected by sequential species loss. The error bars represent the standard deviation of data ranges. Variation explained by two principal coordinate dimensions is given in parentheses by percentage. p values and Sq. root of PERMANOVA are also given. (B) Relative abundance of dominant functional categories at level 1 (mean > 1%) affected by sequential species loss. (C) Heatmaps showing normalized relative abundance of dominant functional categories at level 1 (mean > 1%) affected by sequential species loss. Dendrograms of hierarchical cluster analysis grouping functions are shown. AAD, amino acids and derivatives; CBS, clustering-based subsystems; CDC, cell division and cell cycle; CHO, carbohydrates; CVP, cofactors, vitamins, prosthetic groups, pigments; CWC, cell wall and capsule; DNA, DNA metabolism; FAL, fatty acids, lipids, and isoprenoids; IAM, iron acquisition and metabolism; MAC, metabolism of aromatic compounds; MEM, membrane transport; MIS, miscellaneous; MOT, motility and chemotaxis; NIT, nitrogen metabolism; NUC, nucleosides and nucleotides; PHM, phosphorus metabolism; POT, potassium metabolism; PPT, phages, prophages, transposable elements, plasmids; PRO, protein metabolism; RCS, regulation and cell signaling; RES, respiration; RNA, RNA metabolism; STR, stress response; SUL, sulfur metabolism; and VDD, virulence, disease, and defense.