Figure 1.
Fatty acylated EETI-II molecules are internalized into mammalian cells more efficiently than WT EETI-II
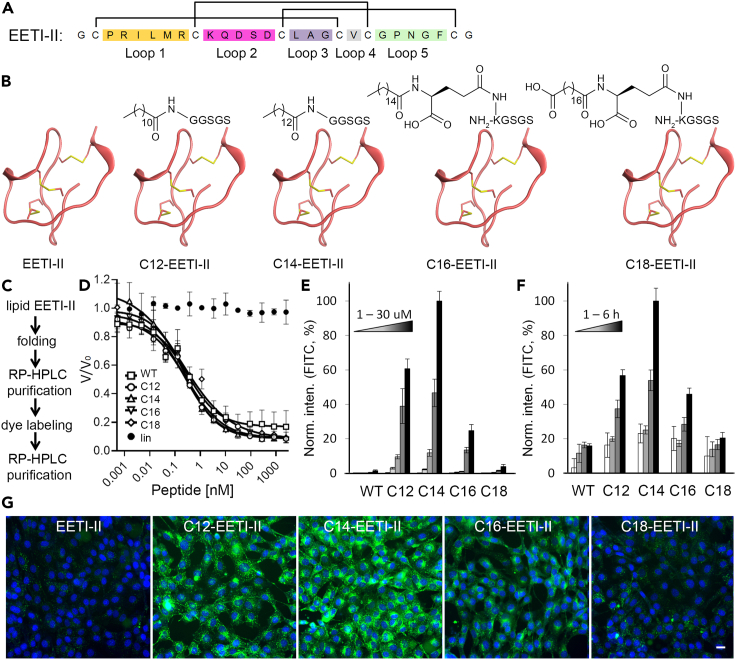
(A) Sequence and disulfide bond connectivity of EETI-II. Each colored region represents a loop region that is flanked by two cysteine residues.
(B) Chemical structures of designed fatty acylated EETI-II peptides.
(C) Strategy for generation of Alexa 488-labeled lipid EETI-II molecules.
(D) Fatty acylated EETI-II peptides show the same trypsin inhibition activity as wild-type (WT) EETI-II. A reduced linear version of EETI-II (lin-EETI) was used as negative control. V/V0 is normalized proteolytic activity.
(E and F) Concentration- and time-dependent uptake of Alexa 488 labeled WT and fatty acylated EETI-II assayed by fluorescence microscopy. NIH3T3 cells were treated with E) increasing concentrations (1, 5, 10, 20, 30 μM) of Alexa 488-labeled WT (EETI-II) or fatty acylated EETI-II for 2 h, or f) 5 μM of Alexa 488-labeled WT or fatty acylated EETI-II for 1, 2, 3, 6 h.
(G) Representative images of cells treated with 20 μM Alexa 488 labeled WT and fatty acylated EETI-II for 2 h. Green: WT or fatty acylated EETI-II; blue: nuclei. Scale bar, 20 μm. Cells were processed as described in methods. Cells were washed with PBS and fixed with 4% PFA. All samples (E–G) were imaged on a high throughput ImageXpress Micro XL imaging system (Molecular Devices) with a 40× objective and images were analyzed by MetaXpress 4.0. Integrated fluorescence intensity values above a threshold defined using the DMSO-treated samples were measured and normalized to samples with the highest signal. Values represent mean ± SD. n = 1,000 cells. Representative images from at least three independent experiments are shown.