Abstract
Biomolecular condensation through phase separation may be a novel mechanism to regulate bacterial processes, including cell division. Previous work revealed that FtsZ, a protein essential for cytokinesis in most bacteria, forms biomolecular condensates with SlmA, a protein that protects the chromosome from damage inflicted by the division machinery in Escherichia coli. The absence of condensates composed solely of FtsZ under the conditions used in that study suggested this mechanism was restricted to nucleoid occlusion by SlmA or to bacteria containing this protein. Here we report that FtsZ alone, under physiologically relevant conditions, can demix into condensates in bulk and when encapsulated in synthetic cell-like systems generated by microfluidics. Condensate assembly depends on FtsZ being in the GDP-bound state and on conditions mimicking the crowded environment of the cytoplasm that promote its oligomerization. Condensates are dynamic and reversibly convert into filaments upon GTP addition. Notably, FtsZ lacking its C-terminal disordered region, a structural element likely to favor biomolecular condensation, also forms condensates, albeit less efficiently. The inherent tendency of FtsZ to form condensates susceptible to modulation by physiological factors, including binding partners, suggests that such mechanisms may play a more general role in bacterial division than initially envisioned.
Keywords: Phase separation, Microfluidics, Macromolecular crowding, Membraneless compartments, Subcellular organization
1. Introduction
Biomolecular condensation is emerging as an important regulatory mechanism involved in the function and subcellular organization of proteins in a wide variety of systems, but its role in bacterial cells has not been appreciated until relatively recently [1]. The dynamic compartments formed through this mechanism, which lack lipid membranes and are in contact with the surroundings from which they physically separate, accumulate specific molecules while excluding others. From an increasing number of studies of mainly eukaryotic proteins, it seems that condensation is favored by macromolecular crowding, protein multivalency and unstructured domains [2,3]. Nucleic acids are often present in these assemblies and they generally enhance the tendency of proteins to form them, probably because of the associated multivalency of the complexes [4–6].
There have been relatively few studies dealing with condensates involving bacterial proteins [1,7–12]. Among them, we recently described condensates [13] involving FtsZ, a key protein whose polymers organize into a dynamic ring-like structure required for bacterial division [14], and SlmA, a DNA-binding protein that blocks FtsZ ring assembly over nucleoids in E. coli through direct interaction with FtsZ [15,16]. On the basis of this finding, we proposed that biomolecular condensation may play a role in the regulation of bacterial division, through the modulation of nucleoid occlusion by SlmA [13].
FtsZ, one of the most conserved proteins across bacterial species, is the central protein of the cytokinesis machinery. From a structural point of view, FtsZ monomers contain a few disordered amino acids at their extreme N-terminus, followed by a globular domain, an unstructured flexible linker (~50 amino acids) and a conserved C-terminal tail [17]. The GDP bound protein forms non-cooperative isodesmic oligomers, through a process dependent on salt and magnesium concentration [18]. These oligomers become larger in physiological crowding conditions [19] and it is under these circumstances when, upon binding to SlmA, FtsZ organizes into dynamic biomolecular condensates, enhanced by DNA carrying the specific SlmA binding sites (SBS; [13]). In the presence of GTP and following a cooperative mechanism [20], FtsZ associates into filaments of different sizes and morphologies that remain assembled until GDP accumulates as a consequence of GTP hydrolysis.
So far, biomolecular condensates formed by FtsZ on its own have not been found in vitro. However, as condensation has been frequently associated with multivalency and unstructured regions, it seems that FtsZ itself would be a good candidate for this type of behavior. Furthermore, the plasticity of FtsZ, reflected in its ability to self-associate into different structures depending on experimental parameters, suggests that FtsZ condensates might form under conditions not previously tested. FtsZ condensates have not been reported in vivo either, although there are evidences of patches and relatively rounded structures of unknown composition, but containing FtsZ, outside and inside the FtsZ-ring [20–22].
Here we have analyzed whether FtsZ itself could undergo biomolecular condensation. We have evaluated the effects of solution conditions, within the physiological ranges described in E. coli [17,23], on the ability of the protein to form condensates, either in bulk or when encapsulated in cytomimetic containers generated by microfluidics. The crowding effect has been also tested with inert polymers typically used in the characterization of these structures, at concentrations mimicking the intracellular conditions. FtsZ condensates were studied by turbidity and fluorescence microscopy, and the impact of removal of the protein unstructured linker on condensation was assessed. This analysis indicates that the main constituent of the bacterial division ring, FtsZ, has the intrinsic ability to form biomolecular condensates, which may be important for the regulation of its function.
2. Results
2.1. GDP bound FtsZ forms micrometer-sized condensates in crowding conditions
We tested the ability of FtsZ to form biomolecular condensates on its own by exploring conditions known to impact its oligomerization tendency, such as magnesium, protein and salt concentrations, and macromolecular crowding. Confocal images of FtsZ labeled with Alexa 488 (FtsZ-Alexa 488) showed that, at 100 mM KCl, 10 mM Mg2+ and in the presence of dextran, Ficoll or PEG as crowders, the protein (5 μM) formed round structures that resembled biomolecular condensates (Fig. 1a). Turbidity measurements on these samples further confirmed the formation of higher order structures in the presence of crowding agents (Fig. 1b), with absorbance values in the absence of crowders virtually zero (0.002 ± 0.002 with 16 μM FtsZ). The observed turbidity trend was compatible with the existence of a threshold protein concentration above which condensates would start forming. Analysis of the data rendered values for this saturation concentration, csat, of 1.3 ± 0.2 μM, 3 ± 1 μM and 3.8 ± 0.1 μM in dextran, Ficoll and PEG, respectively (Fig. 1b). This concentration threshold has been recently established as an additional criterion for identifying phase separation phenomena yielding condensation [3,24].
Fig. 1.
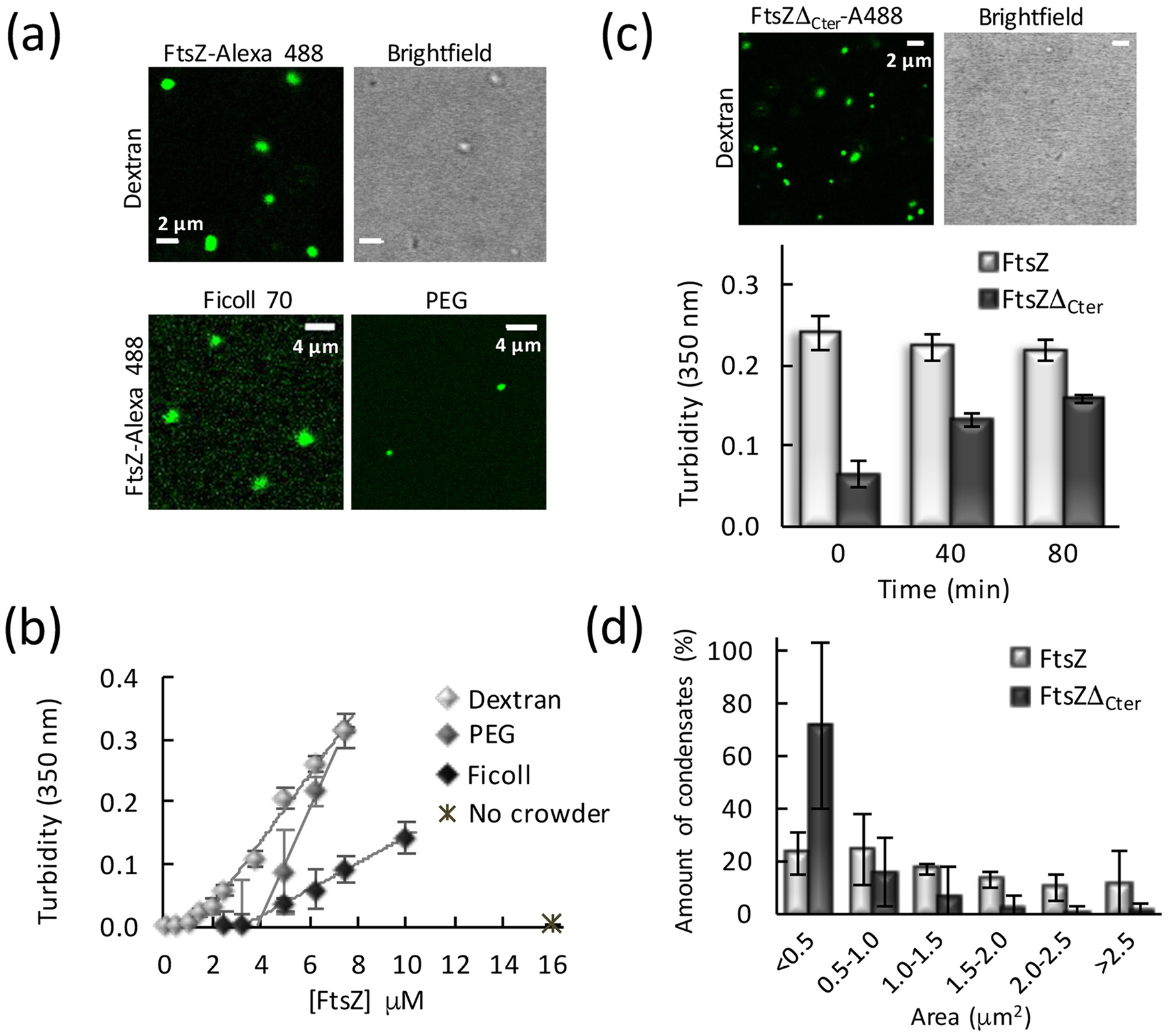
FtsZ forms condensates. (a) Confocal microscopy and transmitted images of condensates of FtsZ in different crowders. (b) Dependence of the turbidity signal of wild-type FtsZ on protein concentration. Lines correspond to a linear model fit to the data. (c) Confocal microscopy and transmitted images of condensates formed by the FtsZ mutant, FtsZΔCter, in dextran. Below, changes in the turbidity signal of the FtsZ mutant condensates over time, with those of wild-type FtsZ shown as reference. (d) Size distribution of FtsZ (n = 99 particles) and FtsZΔCter condensates (n = 238 particles) in the presence of dextran. Errors correspond to S.D. from 3 (FtsZ) or 6 (FtsZΔCter) independent images. Except when specified, FtsZ and FtsZΔCter concentrations were 5 μM. Dextran and Ficoll concentrations were 200 g/l, and PEG was 75 g/l. All experiments in working buffer. In (b) and (c) data are the average of at least 3 independent experiments ± S.D.
Thorough characterization in the presence of dextran showed that the size, shape and abundance of the FtsZ condensates depended on the salt, magnesium and protein concentration. Under the above mentioned Mg2+ and KCl conditions, and up to 5 μM FtsZ, a population of mostly round structures resembling condensates was observed by confocal microscopy (Fig. 1a and Supplementary Fig. S1a). The size distribution of the condensates increased with protein concentration (Fig. S1b). Accordingly, the turbidity values also increased with protein concentration (Fig. 1b). At high protein concentrations, we found a progressive appearance of irregular FtsZ assemblies of large size at the expense of the smaller and more regular structures (Supplementary Fig. S1a). Regarding the effect of Mg2+, lowering its concentration to 1–5 mM significantly reduced the number of structures, except if protein concentration was tripled to 16 μM, as determined through confocal microscopy and turbidity measurements (Supplementary Fig. S1a,c). Lastly, an increase in salt to 300 mM KCl resulted in low turbidity values, consistent with scarce and very small structures, even with 10 mM Mg2+ (Supplementary Fig. S1d,e). Only at high protein concentrations was a significant population of arrangements of variable shape apparent (Supplementary Fig. S1e). These results are in line with our previous study on FtsZ⋅SlmA⋅SBS biomolecular condensates, at 1 mM Mg2+, where FtsZ alone was homogeneously dispersed [13]. Condensation in the presence of Ficoll followed the same trend with KCl and Mg2+ concentration as that described with dextran (Supplementary Fig. S2a, b).
These experiments show the formation of FtsZ round structures, above a saturation concentration of protein, under salt and ionic strength conditions rendering sufficient multivalency through the promotion of the protein monomers self-association. Regulation of the assembly of these defined micrometer-sized arrangements by ion concentrations and salt suggests an implication of electrostatic forces in their formation, without excluding other possible types of interactions. As round structures compatible with biomolecular condensates prevailed at 5 μM FtsZ, in 50 mM Tris-HCl, pH 7.5 with 100 mM KCl, 10 mM MgCl2 (working buffer) and 200 g/l dextran, these conditions were selected for their further characterization.
2.2. Condensation of FtsZ lacking the unstructured C-terminal region is less efficient
Unstructured domains of proteins have been implicated as important contributors to biomolecular condensation. To evaluate the impact of such domains on the formation of the condensate-like arrangements described above, we investigated the mutant FtsZΔ315–383 (FtsZΔCter), which contains the globular domain of FtsZ but lacks the unstructured C-terminal flexible linker as well as the conserved C-terminal peptide known to bind other cell division proteins [25]. We found that this mutant FtsZ retained the ability to form condensates in crowded solutions (Fig. 1c). Interestingly, these condensates in the images were substantially smaller than those formed by the wild-type protein when visualized at short times after their formation, also reflected in the different size distribution of the condensates of both proteins at the same concentration (Fig. 1d). This observation is compatible with the lower turbidity values measured for the mutant protein after mixing with the crowder. While the turbidity values remained constant for at least 80 min in the case of the wild-type FtsZ, those for the mutant noticeably increased within this same time interval, suggesting that the condensation process is slower (Fig. 1c and Supplementary Fig. S3a). Notably, evaluation of the dependence with mutant concentration of the turbidity signal at equilibrium rendered a csat value similar to that of wild-type, 1.1 ± 0.2 μM (Supplementary Fig. S3b). Analysis of this mutant by analytical ultracentrifugation showed that it retains the ability to form oligomers in solution (Supplementary Fig. S4), which would provide the multivalent states that typically favor formation of condensates. Together, this evidence suggests that the unstructured domain of FtsZ may not be strictly required to mediate its assembly as it did not preclude the formation of these structures, but the delay in condensation indicates it probably has a role in enhancing the overall process.
2.3. FtsZ condensates are dynamic, reversible and evolve into filaments in the presence of GTP
Next, we asked whether the round FtsZ structures resembling molecular condensates were dynamic and reversible. To this end, we followed four different approaches: incorporation of added FtsZ on preformed condensates, evolution of condensates over time, GTP-triggered polymerization of FtsZ from condensates and subsequent reassembly into condensates upon GTP depletion, and dissociation of condensates by salt shift.
First, we performed capture experiments using two separate pools of FtsZ labeled with spectrally different dyes, and similarly to that described for other condensates [13,26]. We found that FtsZ-Alexa 488 readily incorporated into preformed condensates containing FtsZ labeled with Alexa 647 (FtsZ-Alexa 647) as a tracer, suggesting that the population of FtsZ within the condensates is dynamic. Images show the initial and final states and the stepwise diffusion of the added protein into the condensates (Fig. 2a). Capture experiments using the reverse dyes yielded the same result (Supplementary Fig. S5a). Likewise, FtsZΔCter condensates displayed dynamic behavior following the same approach (Fig. 2b).
Fig. 2.
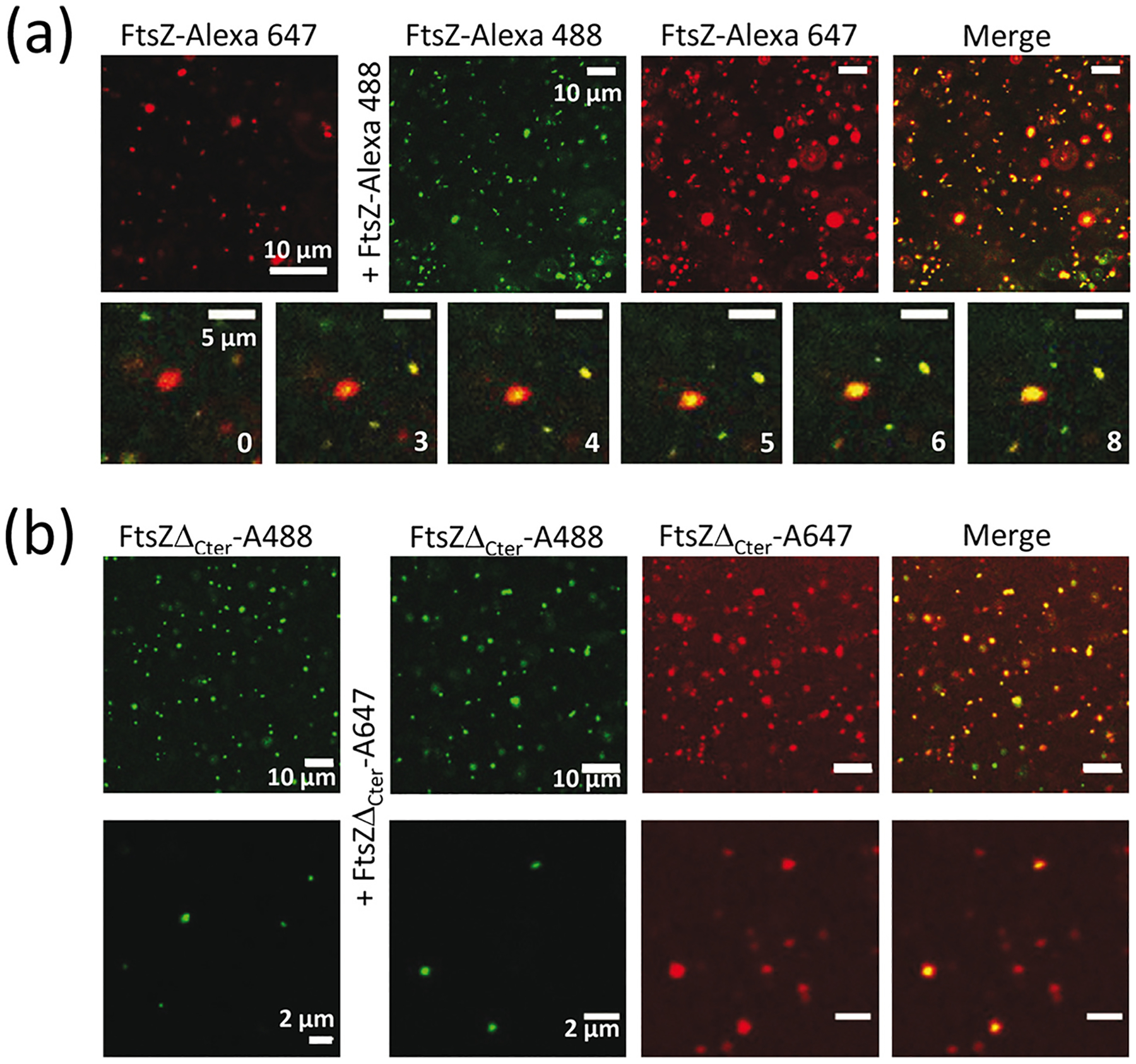
FtsZ condensates are dynamic. (a) Representative confocal microscopy images showing the initial and final states after addition of FtsZ-Alexa 488 into FtsZ condensates with FtsZ-Alexa 647 as a tracer and, below, stepwise diffusion at the times indicated in seconds. (b) Images showing the initial and final states after addition of FtsZΔCter-A647 into mutant condensates with FtsZΔCter-A488 as a tracer. FtsZ and mutant concentrations were 5 μM. All experiments were conducted in working buffer with 200 g/l dextran.
We also monitored the evolution of FtsZ condensates over time. We found a clear shift of the condensate size distribution towards larger values over a 4 h time course (Fig. 3). Growth of the condensates may be the result of fusion, Ostwald ripening, which seems to be a property of cytoskeletal proteins [27], or a combination of both. Capture experiments conducted on these grown condensates revealed they remain dynamic in terms of protein incorporation (Supplementary Fig. S5b).
Fig. 3.
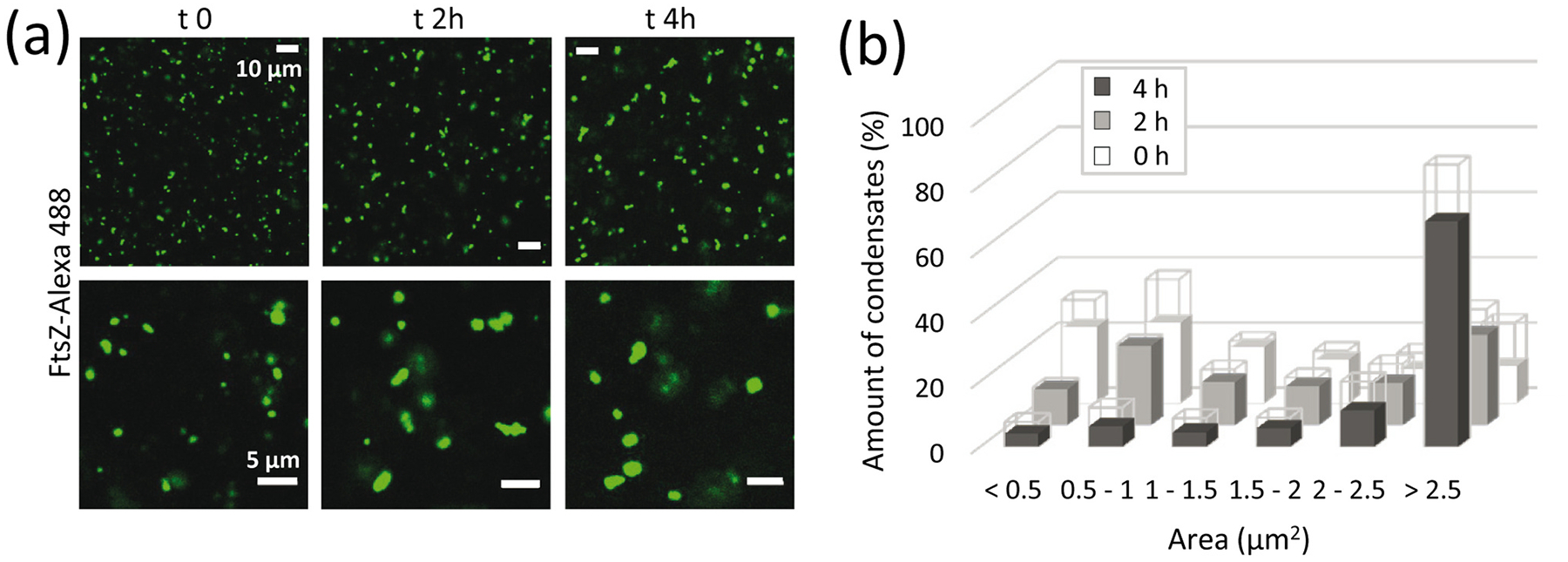
FtsZ condensates grow with time. (a) Images of FtsZ condensates with time, and (b) corresponding distribution of sizes (n = 99, 157 and 206 particles for 0, 2 and 4 h, respectively). Errors, depicted as open sections of the bars, correspond to S.D. from 2 (2 h) or 3 (0 and 4 h) independent images. FtsZ concentrations were 5 μM. All experiments were conducted in working buffer with 200 g/l dextran.
Next, we tested the responsiveness of the FtsZ condensates to GTP, which is well known to induce FtsZ polymer formation and is a hallmark of its functionality in vivo. Confocal images showed that addition of GTP strongly induced the polymerization of FtsZ that seemed to occur with a concomitant reduction in the number and/or size of the condensates, although both structures coexisted during the time window monitored (Fig. 4a). Turbidity measurements showed a significant decrease in the signal arising from the condensates shortly after GTP addition (Fig. 4b and Supplementary Fig. S6a). Moreover, measurements with condensates formed using Ficoll as crowding agent exhibited the same behavior (inset in Supplementary Fig. S6a). This, together with the incipient polymer formation observed in the images taken at short times, strongly suggests that the polymers originate from the condensates. Had polymerization exclusively come from the unassembled protein coexisting with condensates, we would have detected an increase in turbidity instead of the strong decrease observed. Monitoring these samples over longer times showed that the turbidity signal continued to slightly decrease up to a point, dependent on the nucleotide concentration, above which it gradually recovered, reaching a value close to that in the absence of GTP (Fig. 4b and Supplementary Fig. S6b). This behavior is consistent with the characteristic dissociation of the FtsZ polymers due to GTP exhaustion [28]. Interestingly, the round condensates after four hours also exhibited conversion into the typical GTP-triggered FtsZ polymers, as monitored by confocal microscopy (Supplementary Fig. S6c).
Fig. 4.
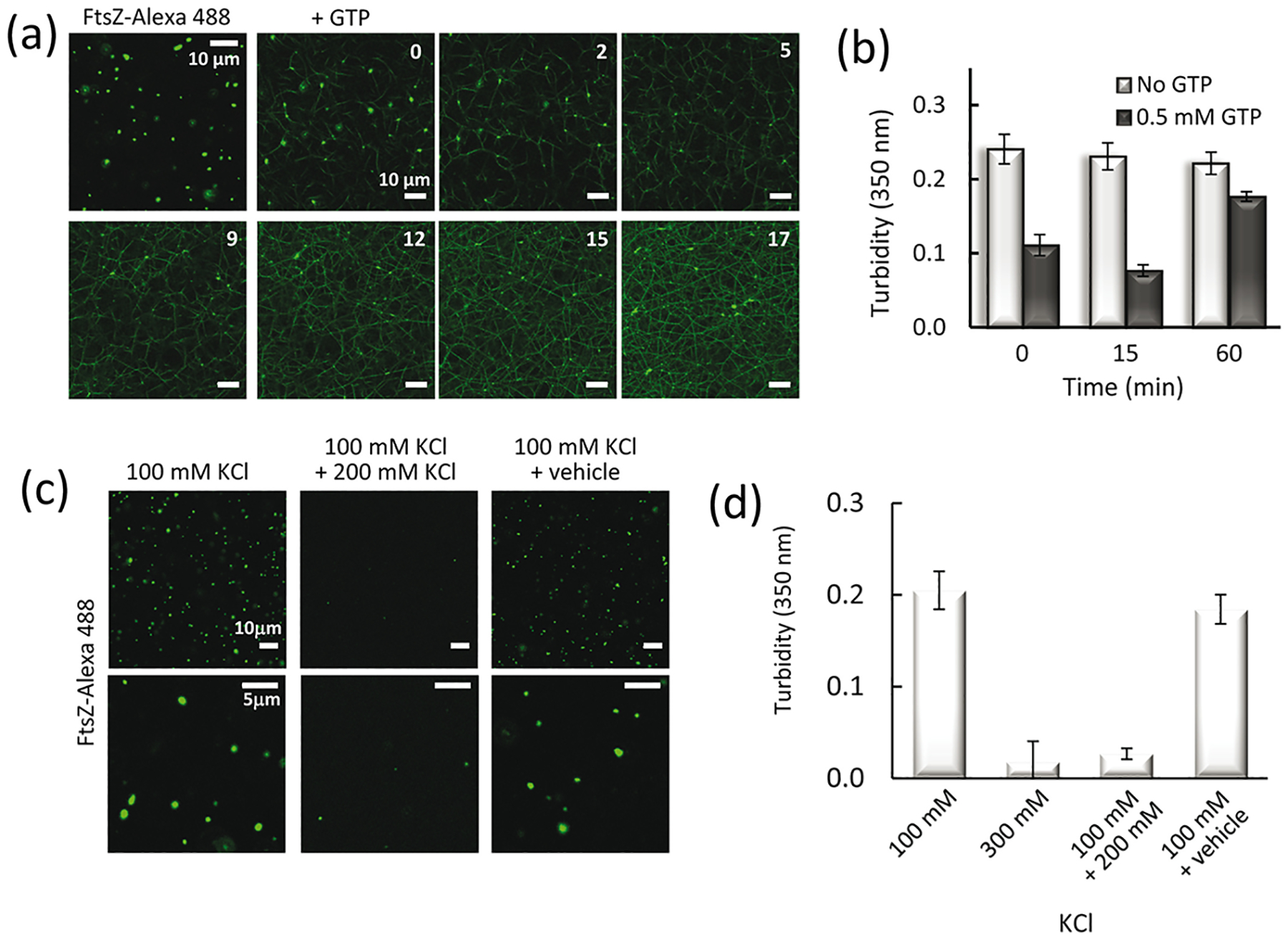
Condensates are reversible and FtsZ within them remains active for polymerization. (a) Representative images of FtsZ condensates before polymerization and evolution after GTP (0.5 mM) addition at the indicated times, in minutes. All images of the time lapse correspond to a single field. (b) Evolution of the turbidity signal of condensates with time and variation after GTP addition at time zero. (c) Images showing FtsZ condensates and their dissociation by addition of KCl up to 300 mM. Images on the far right show the effect of the addition of vehicle instead (the same volume of water without KCl) to account for possible dilution effects (7%). (d) Turbidity values corresponding to the samples in (c). The turbidity of a sample directly prepared in 300 mM KCl is shown for comparison. In (b) and (d), values correspond to the average of 3 independent experiments ± S.D. FtsZ concentration was 5 μM. All experiments were conducted in working buffer with 200 g/l dextran.
Lastly, the reversible nature of FtsZ condensates was assessed by measuring the ability of the protein to assemble into polymers, after addition of 0.1 mM GTP to condensates formed under the working conditions, and back to condensates upon GTP depletion. As shown by confocal microscopy, upon depolymerization the protein formed condensates similar to those before nucleotide addition (Supplementary Fig. S6d). Accordingly, and as observed in the presence of other GTP concentrations, the initial decrease in the turbidity signal was followed by a recovery (Supplementary Fig. S6b). Further evidence of the reversibility of FtsZ condensates was obtained by inducing a salt shift on condensates formed at these working conditions (i.e. 100 mM KCl). The increase up to 300 mM KCl showed a dramatic reduction in the size and number of condensates, as observed by confocal microscopy (Fig. 4c) accompanied by a drop in the turbidity signal values, matching those of the condensates originally formed at this higher salt concentration (Fig. 4d).
2.4. Reconstruction of FtsZ condensates in cytomimetic microdroplets
To determine whether FtsZ condensates could also be formed in a confined cell-like system, we used microfluidics to encapsulate the protein (Fig. 5). Confocal microscopy images of FtsZ, containing FtsZ-Alexa 488 as a tracer, encapsulated in microdroplets stabilized by an E. coli lipid mixture boundary, indicated the presence of condensates in the z-sections of the microdroplets with no apparent preference for the lumen or the lipid interface. The presence of these structures was more obvious in the maximal projection of the images (far right in Fig. 5a) that evidenced their presence in all microdroplets. No background signal was observed in the lumen, while a strong fluorescence demarcating the lipid boundary was found.
Fig. 5.
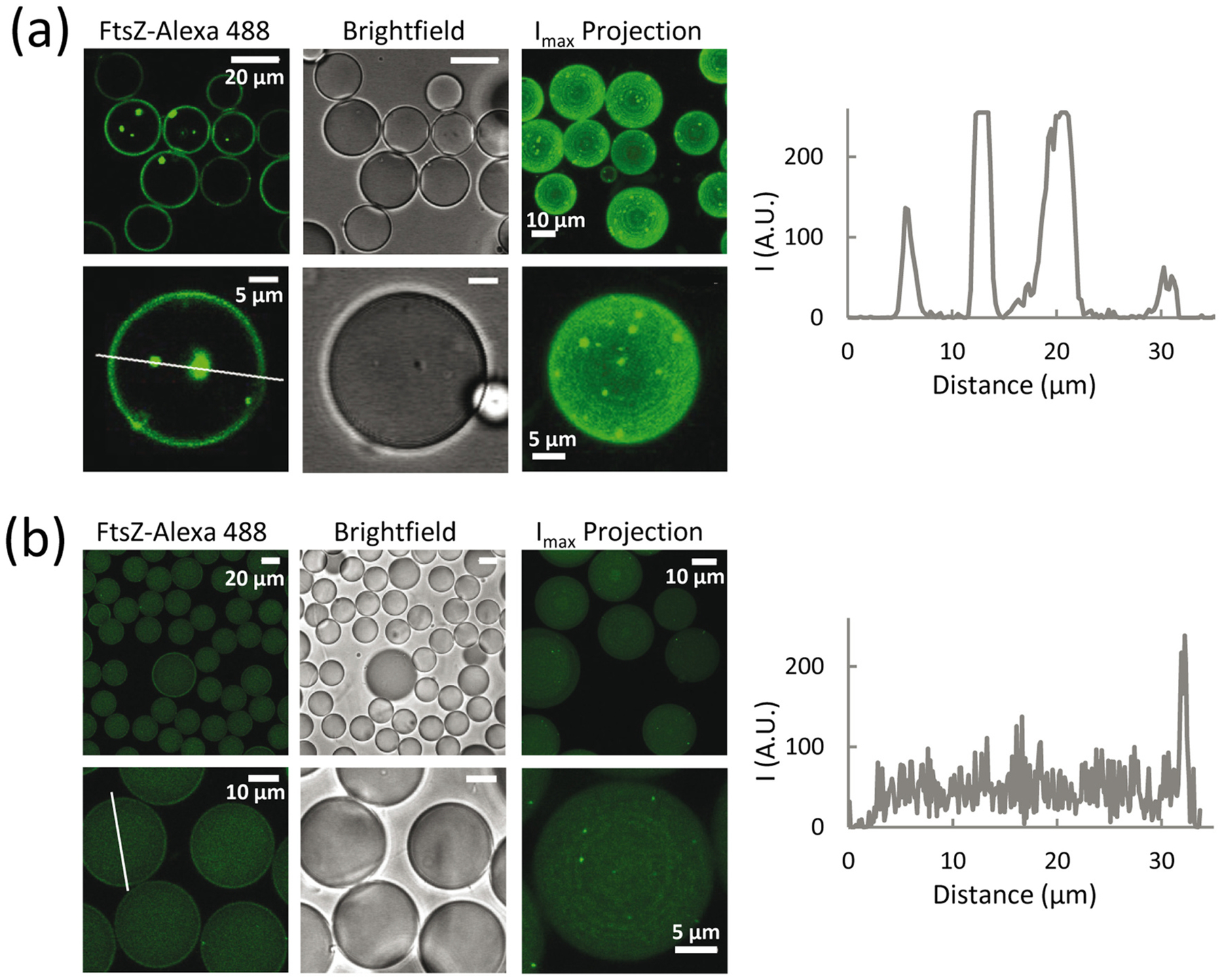
FtsZ condensates are also formed in confined cell-like systems. (a) Confocal microscopy images of microfluidics microdroplets stabilized by E. coli lipids and containing the condensates. 100% of the microdroplets examined (n = 40) contained condensates. Experiments were with 5 μM FtsZ in working buffer with 200 g/l dextran. (b) Encapsulation under experimental conditions not promoting FtsZ condensation in bulk: 12 μM FtsZ in 50 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 300 mM KCl and 150 g/l dextran. 58% of the microdroplets (n = 40) contained condensates. The third column of images for (a) and (b) are maximum intensity projections corresponding to different fields. To the right of (a) and (b) are intensity profiles of the green channel obtained across the lines drawn in the respective images.
Encapsulation of FtsZ under conditions that discourage bulk formation of condensates or any other detectable structures (300 mM KCl, 1 mM MgCl2, 150 g/l dextran) showed scarce and very small condensates in the z-sections of 58% of the microdroplets tested, which were more obvious in the maximal projection and mostly near the membrane (Fig. 5b). It should be noted that the percentage of microdroplets containing condensates under these conditions might be underestimated by the inability to detect condensates smaller than the resolution of confocal microscopy. Under these conditions, the protein remained homogeneously distributed within the lumen, in good agreement with previous reports showing FtsZ encapsulated under similar conditions [29,30].
These experiments confirm that condensates are still assembled when FtsZ is encapsulated under conditions favoring their formation in bulk. Moreover, incipient condensate formation was also apparent when the protein was encapsulated in conditions under which no condensates were detected by confocal microscopy or turbidity, strongly suggesting that confinement and/or the lipid membrane, directly or indirectly, influence their formation.
3. Discussion
Here we show that purified FtsZ protein forms reversible structures compatible with biomolecular condensates, observed under conditions resembling the physiological ones both in bulk solutions and when reconstructed in cytomimetic platforms. These structures are mostly reservoirs of material that have exceeded their solubility in the phase outside the condensate. Their assembly, disassembly, abundance, and coexistence (when the protein concentration is increased to high levels) with irregular clusters are all influenced by crowding and other chemical conditions that affect oligomerization of GDP-bound FtsZ (Fig. 6a). This behavior is probably derived from the well-known self-association properties of this protein, which provide the multivalent states that typically favor formation of condensates. The present work emphasizes the key role of GDP in the formation of FtsZ condensates, different from that in other NTP-bound proteins such as DEAD-box ATPases where ATP hydrolysis promotes disassembly of the condensates [31].
Fig. 6.
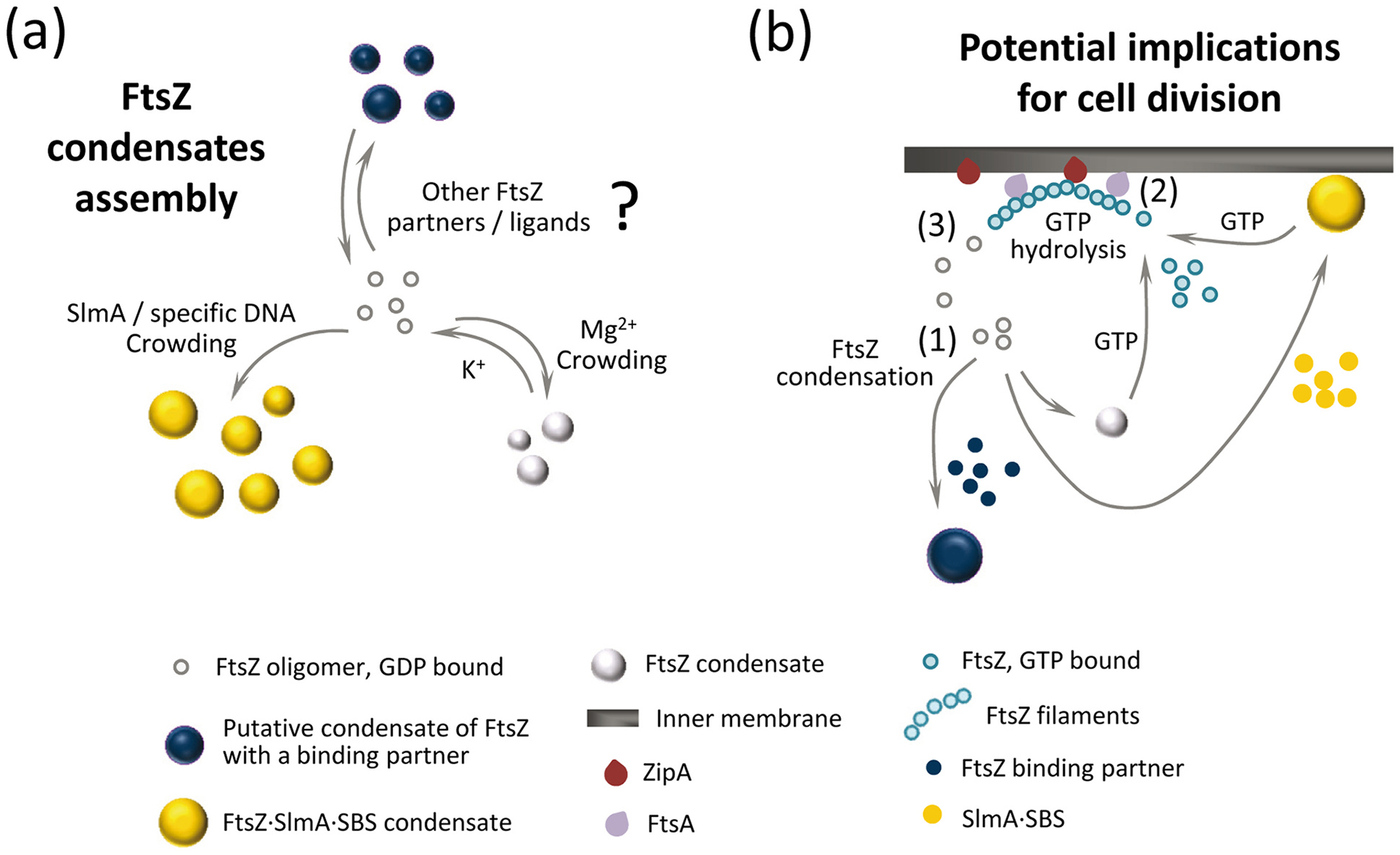
Formation of FtsZ biomolecular condensates and potential role in bacterial cell division regulation. (a) FtsZ condensate assembly is disfavored by K+, favoured by crowding and Mg2+ and strongly promoted by the SlmA⋅SBS nucleoprotein complex. It is possible that condensation is also positively or negatively regulated by additional binding partners or by natural or synthetic ligands of FtsZ. (b) In the cell, FtsZ oligomers could form condensates on their own or together with the nucleoid occlusion factor SlmA (FtsZ⋅SlmA complexes tend to locate at the cellular membrane [13]), and possibly also with other division regulators (1). In the presence of GTP, FtsZ would leave the condensates and associate into filaments attached to the bacterial membrane through the natural anchors, ZipA and FtsA (2). GTP hydrolysis would lead to GDP-bound FtsZ subunits within the filaments, and a loss of their longitudinal interactions would result in the release of FtsZ (3). The released FtsZ would be able to reassemble into homotypic or heterotypic dynamic condensates, contributing to the spatiotemporal regulation of bacterial cell division.
FtsZ within the condensates remained responsive to ligands, only forming filaments upon addition of GTP. This GTP-dependent shuttling of FtsZ between condensate and polymer states was also observed in our previous study of FtsZ⋅SlmA complexes, which showed that when mixed with the DNA-binding SlmA protein under crowding conditions, FtsZ formed condensates capable of reversible evolution to polymers in the presence of GTP [13]. Condensation seemed more favored in that case, as such structures were observed under a substantially wider range of conditions in which FtsZ on its own was not able to form condensates. This relative promiscuity could be explained by the higher multivalency conferred on the system by the presence of SlmA, a dimer that binds in pairs to a specific SBS site on DNA [32], while also establishing contacts with the C-terminal tail and the folded domain of FtsZ [33].
Protein condensate formation is usually associated with regions of low sequence complexity [2] and, indeed, FtsZ contains an unstructured segment [34]. The mutant FtsZ, deprived of this region, shows condensation possibly related to the multivalency emerging from its oligomerization interfaces. Nonetheless, this mutant displays slower condensation than wild-type FtsZ, which might be a result of differences in their dynamics and/or topology. Along these lines, GTP-induced filaments of FtsZ mutants from different bacterial species lacking the C-terminal peptide and/or the linker exhibited morphological alterations and slower polymerization rates [35]. Also, as here with FtsZ, examples can be found where condensation is observed for folded domains, with disordered regions playing a modulatory role [9,36–38].
We observed formation of FtsZ condensates in confined cell like environments that included crowding and a membrane boundary. The use of such synthetic systems with controlled composition simplifies the interpretation of condensation compared with in vivo studies and, in the particular case of bacteria, circumvents technical obstacles due to their small size. Our encapsulation studies indicate that fully formed FtsZ condensates do not show a preference for the membrane, although it seems that the membrane could participate in the condensation of FtsZ, as shown by the small emerging structures at the lipid surface under bulk non-condensation conditions. This is compatible with either an increase in size of sub-micrometer condensates already present in solution, providing a surface for nucleation [39], and/or further enhancement of their formation by confinement within the microdroplet. Interestingly, FtsZ⋅SlmA nucleoprotein condensates do accumulate at the lipid boundary [13], probably because of SlmA, which we recently found to interact with membranes [40].
How might biomolecular condensation of FtsZ be relevant in vivo? Under certain conditions, cellular FtsZ oligomers might assemble into condensates, either alone or combined with any of FtsZ’s multiple binding partners (Fig. 6b). Condensate formation would be modulated to some extent by weak, transient electrostatic interactions with the environment, as nucleic acids are abundant and most proteins in E. coli are polyanions at intracellular pH [41]. As condensation enables dynamic spatial localization of cellular processes, condensates containing FtsZ might be favored during a particular phase of the cell cycle, perhaps used as a cytoplasmic storage form, or as a response to stresses (see below). For instance, the local accumulation of a higher number of molecules provided by condensation could permit more rapid preassembly with key protein partners or ligands than by depending on successive recruitment of cell division proteins in the cytoplasm or on the cytoplasmic membrane. In the presence of GTP, FtsZ would exit the condensates and associate into mobile complexes of filaments attached to the bacterial membrane through the natural anchor proteins, ZipA and FtsA [22,42,43]. GTP hydrolysis would increase the amount of FtsZ subunits bound to GDP within the filaments, with the associated loss in their longitudinal interactions and subsequent release of FtsZ [20], now available for a new condensation cycle. Curiously enough, the elusive ultrastructure of the E. coli Z-ring, only recently revealed through high resolution imaging, shows loosely associated clusters of FtsZ molecules of round appearance [20,21]. Further work will be required to ascertain the interplay of the FtsZ condensates identified here or of other putative heterotypic condensates involving this protein with the division ring.
The biomolecular condensation of purified FtsZ demonstrated here adds to the growing number of examples of this type of behavior by bacterial proteins. Despite initial doubts raised by the lower frequency of intrinsically disordered regions in bacterial proteomes, very recent research suggests that biomolecular condensation is an organizational principle that also operates in these microorganisms [1]. As an alternative to the membranous organelles of eukaryotes, such condensates distributed within the bacterial cytoplasm might contribute to the spatial regulation of various metabolic reactions, a role once attributed almost exclusively to the bacterial membrane envelope. In particular, condensates might form preferentially under stress conditions. Interestingly, it was recently reported that late stationary phase E. coli assemble protein aggregates at their cell poles containing FtsZ and other proteins; these “regrowth delay bodies” dissolve once growth is resumed, suggesting that they are dynamic [44]. Such bodies may consist of multiple proteins that tend to form condensates under certain stress conditions such as starvation, desiccation and persister states [45]. It is noteworthy that cells in such states are likely deficient in GTP, a situation that as suggested by our results, could contribute to the formation of FtsZ condensates. Given that FtsZ is considered a promising target in the quest for new antibiotics [46], and that cells in the persister state are particularly resistant to antibiotics [47], understanding how FtsZ forms condensates and where and when such condensates might form in cells may provide additional clues to fight against antimicrobial resistance.
4. Materials and methods
4.1. Reagents
GTP, dextran 500 (500 kDa), PEG 8 (8 kDa) and other analytical grade chemicals were from Sigma Chemical Co., St. Louis MO, USA. Ficoll 70 (70 kDa) was from GE Healthcare, IL, USA. Crowders were dialyzed in 50 mM Tris-HCl, 100 or 300 mM KCl, pH 7.5, and their concentration measured as earlier described [48]. Polar extract phospholipids from E. coli (Avanti Polar Lipids, Alabaster AL, USA), were stored in chloroform at −20 °C. Shortly before use they were thoroughly dried in a Speed-Vac device and the resulting film resuspended in mineral oil by two cycles of vortex and 15 min sonication in a bath. Final concentration of the lipids in mineral oil was 20 g/l.
4.2. FtsZ and FtsZΔCter expression, purification and labeling
Wild-type E. coli FtsZ was isolated as described elsewhere [18] and stored at −80 °C until used. Protein obtained through this protocol is bound to GDP. The plasmid expressing the FtsZΔCter mutant (FtsZΔ315–383, which lacks residues comprising the unstructured region and the C-terminal tail of FtsZ) was kindly provided by P. Schwille (Max Planck Institute of Biochemistry, Martinsried) and purified following the same procedure. Covalent labeling of amine groups of the proteins with Alexa Fluor 488 or Alexa Fluor 647 succinimidyl ester dyes (Molecular probes/Invitrogen) was conducted in its polymer-assembled form as previously described [49,50]. The labeling ratio, calculated from the molar absorption coefficients of the proteins and the dyes, ranged between 0.3 and 0.6 (wild type) and 0.1–0.2 (mutant) moles of dye per mole of protein.
4.3. Preparation of bulk samples and selection of final conditions
FtsZ was directly added to solutions containing the crowders at the specified conditions of KCl and magnesium in 50 mM Tris-HCl, pH 7.5, and incubated for ~30 min, unless otherwise indicated. Except when stated, FtsZ was bound to GDP, with the nucleotide coming from the purification protocol employed (see 4.2). When required, polymerization was triggered by diffusion of GTP directly added over the mixture. For imaging experiments, labeled proteins were used as tracers (final concentration 0.5 or 1 μM, <10% of total protein concentration). Images were acquired with either Alexa 488 or Alexa 647 dyes, both for wild-type (FtsZ-Alexa 488, FtsZ-Alexa 647) and mutant proteins (FtsZΔCter-A488, FtsZΔCter-A647) with equivalent results. Unless otherwise specified, experiments were conducted with 5 μM FtsZ in working buffer (50 mM Tris-HCl pH 7.5, with 100 mM KCl, 10 mM MgCl2) and 200 g/l dextran.
4.4. Microfluidic chip fabrication and FtsZ encapsulation
The devices used were constructed by conventional soft lithographic techniques from masters kindly provided by the W.T.S. Huck group (Radboud University, Nijmegen, The Netherlands; chip design detailed elsewhere [29]) following the procedure described [13].
Encapsulation was conducted by mixing, in a 1:1 ratio prior to the droplet formation junction, two streams of dispersed aqueous phases containing FtsZ-Alexa 488 (0.5 μM) as a tracer and, except when stated, FtsZ (5 μM) in working buffer with 200 g/l dextran. The third stream supplied the E. coli lipid mixture at 20 g/l in mineral oil. Data presented correspond to experiments delivering solutions at 150 μl/h (oil phase) and 20 μl/h (total aqueous phases) by automated syringe pumps (Cetoni GmbH, Germany) yielding uniform droplets with average diameters of 22 μm. Production of the lipid microdroplets in the microfluidic chip was monitored using an Axiovert 135 fluorescence microscope (Zeiss).
4.5. Diffusion of additional FtsZ into preformed condensates
Samples with FtsZ at the specified final concentration containing 0.5 μM FtsZ labeled with Alexa 647 as a tracer were prepared and imaged before and after addition of 0.5 μM FtsZ-Alexa 488. Experiments adding FtsZ-Alexa 647 to condensates labeled with FtsZ-Alexa 488 as a tracer were also conducted. The diffusion of the added labeled protein into the condensates was monitored over time. Experiments with the mutant were done following the same procedure.
4.6. Confocal microscopy measurements and data analysis
Condensates and microfluidics microdroplets were visualized in silicone chambers (Molecular probes/Invitrogen) glued to coverslips. Images were obtained with Leica TCS-SP2 or TCS-SP5 inverted confocal microscopes with a HCX PL APO 63× oil immersion objective (N.A. = 1.4; Leica, Mannheim, Germany). 488 and 633 nm laser excitation lines were used to excite Alexa 488 and Alexa 647 dyes, respectively. Several images were registered across each sample, corresponding to different observation fields. Brightfield and fluorescence images were taken simultaneously. ImageJ (National Institutes of Health, USA) was used to produce images and, after noise reduction by applying a Kuwahara filter and threshold correction by visual inspection, to measure the areas distribution of FtsZ condensates in the confocal images with the particle analysis option of the software. Quantification of microdroplets containing condensates was conducted on microdroplets that had been imaged with a complete z-stack, as condensates can be found anywhere within their volume and a single confocal plane could easily lead to underestimation.
4.7. Turbidity measurements and determination of csat
Turbidity of samples (125 μl solutions) aimed at determining the effect of crowders, salts or FtsZ concentration on condensate formation, and their response to GTP addition, was recorded at room temperature and 350 nm using a Varioskan Flash plate reader (Thermo Fisher Scientific, MA, USA). The absorbance was measured after 30 min incubation. For time dependence measurements, data were taken every 5 min. Reported values are the average of at least 3 independent experiments ± S.D., unless otherwise stated.
The protein concentration above which the condensates form, csat, was determined from the dependence of the turbidity signal with protein concentration. Data scaling with the protein concentration was analyzed by fitting a linear model, rendering a csat value corresponding to the x intercept.
Supplementary Material
Acknowledgments
We thank H. Yébenes for advice on, and preliminary, image analysis and M. Sobrinos-Sanguino for technical support. We also thank the staff of CIB Margarita Salas Confocal Laser and Multidimensional Microscopy (M.T. Seisdedos and G. Elvira) and Molecular Interactions (J.R. Luque) Facilities for excellent assistance in imaging and ultracentrifugation experiments, and the Technical Support Facility for invaluable input. This work was supported by the Spanish Ministerio de Economía y Competitividad (BFU2014-52070-C2-2-P and BFU2016-75471-C2-1-P, AEI/FEDER, UE, to G.R.), by the Spanish Ministerio de Ciencia e Innovación (2019AEP088 and PID2019-104544GB-100/AEI/10.13039/501100011033, to G.R. and S.Z.), and by the National Institutes of Health (GM131705, to W.M.). M.Á.R.-R. was supported by the Agencia Estatal de Investigación and the European Social Fund through grant BES-2017-082003. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Declaration of competing interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbamcr.2021.118986.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding authors upon reasonable request.
References
- [1].Azaldegui CA, Vecchiarelli AG, Biteen JS, The emergence of phase separation as an organizing principle in bacteria, Biophys. J 119 (2020) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry, Nat Rev Mol Cell Biol 18 (2017) 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shin Y, Brangwynne CP, Liquid phase condensation in cell physiology and disease, Science 357 (2017). [DOI] [PubMed] [Google Scholar]
- [4].Lin Y, Protter DS, Rosen MK, Parker R, Formation and maturation of phase-separated liquid droplets by RNA-binding proteins, Mol. Cell 60 (2015) 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP, Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization, Cell 163 (2015) 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M, Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein tau, Nat. Commun 8 (2017) 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Al-Husini N, Tomares DT, Bitar O, Childers WS, Schrader JM, Alpha-Proteobacterial RNA Degradosomes Assemble Liquid-Liquid Phase-Separated RNP Bodies, Mol Cell 71 (2018), 1027–1039 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abbondanzieri EA, Meyer AS, More than just a phase: the search for membraneless organelles in the bacterial cytoplasm, Curr. Genet 65 (2019) 691–694. [DOI] [PubMed] [Google Scholar]
- [9].Wang H, Yan X, Aigner H, Bracher A, Nguyen ND, Hee WY, Long BM, Price GD, Hartl FU, Hayer-Hartl M, Rubisco condensate formation by CcmM in beta-carboxysome biogenesis, Nature 566 (2019) 131–135. [DOI] [PubMed] [Google Scholar]
- [10].Heinkel F, Abraham L, Ko M, Chao J, Bach H, Hui LT, Li H, Zhu M, Ling YM, Rogalski JC, Scurll J, Bui JM, Mayor T, Gold MR, Chou KC, Av-Gay Y, McIntosh LP, Gsponer J, Phase separation and clustering of an ABC transporter in mycobacterium tuberculosis, Proc. Natl. Acad. Sci. U. S. A 116 (2019) 16326–16331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ladouceur AM, Parmar BS, Biedzinski S, Wall J, Tope SG, Cohn D, Kim A, Soubry N, Reyes-Lamothe R, Weber SC, Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation, Proc. Natl. Acad. Sci. U. S. A 117 (2020) 18540–18549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Guilhas B, Walter JC, Rech J, David G, Walliser NO, Palmeri J, Mathieu-Demaziere C, Parmeggiani A, Bouet JY, Le Gall A, Nollmann M, ATP-driven separation of liquid phase condensates in bacteria, Mol. Cell 79 (2020), 293–303 e4. [DOI] [PubMed] [Google Scholar]
- [13].Monterroso B, Zorrilla S, Sobrinos-Sanguino M, Robles-Ramos MA, Lopez-Alvarez M, Margolin W, Keating CD, Rivas G, Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA, EMBO Rep. 20 (2019), e45946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Haeusser DP, Margolin W, Splitsville: structural and functional insights into the dynamic bacterial Z ring, Nat Rev Microbiol 14 (2016) 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schumacher MA, Bacterial nucleoid occlusion: multiple mechanisms for preventing chromosome bisection during cell division, Subcell Biochem 84 (2017) 267–298. [DOI] [PubMed] [Google Scholar]
- [16].Mannik J, Bailey MW, Spatial coordination between chromosomes and cell division proteins in Escherichia coli, Front. Microbiol 6 (2015) 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Erickson HP, Anderson DE, Osawa M, FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one, Microbiol. Mol. Biol. Rev 74 (2010) 504–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rivas G, López A, Mingorance J, Ferrándiz MJ, Zorrilla S, Minton AP, Vicente M, Andreu JM, Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer. The primary steps for FtsZ assembly, J Biol Chem 275 (2000) 11740–11749. [DOI] [PubMed] [Google Scholar]
- [19].Rivas G, Fernandez JA, Minton AP, Direct observation of the enhancement of noncooperative protein self-assembly by macromolecular crowding: indefinite linear self-association of bacterial cell division protein FtsZ, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 3150–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Du S, Lutkenhaus J, At the heart of bacterial cytokinesis: the Z ring, Trends Microbiol. 27 (2019) 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lyu Z, Coltharp C, Yang X, Xiao J, Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional Superresolution imaging, Biopolymers 105 (2016) 725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rowlett VW, Margolin W, 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells, Biophys. J 107 (2014) L17–L20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Record MT Jr., Courtenay ES, Cayley S, Guttman HJ, Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments, Trends Biochem. Sci 23 (1998) 190–194. [DOI] [PubMed] [Google Scholar]
- [24].Peng A, Weber SC, Evidence for and against liquid-liquid phase separation in the nucleus, Noncoding RNA 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ortiz C, Natale P, Cueto L, Vicente M, The keepers of the ring: regulators of FtsZ assembly, FEMS Microbiol. Rev 40 (2016) 57–67. [DOI] [PubMed] [Google Scholar]
- [26].Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, Hyman AA, The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin, Cell 169 (2017), 1066–1077 e10. [DOI] [PubMed] [Google Scholar]
- [27].Wiegand T, Hyman AA, Drops and fibers - how biomolecular condensates and cytoskeletal filaments influence each other, Emerg Top Life Sci. 4 (2020) 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mukherjee A, Lutkenhaus J, Dynamic assembly of FtsZ regulated by GTP hydrolysis, EMBO J. 17 (1998) 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mellouli S, Monterroso B, Vutukuri HR, te Brinke E, Chokkalingam V, Rivas G, Huck WTS, Self-organization of the bacterial cell-division protein FtsZ in confined environments, Soft Matter 9 (2013) 10493–10500. [Google Scholar]
- [30].Sobrinos-Sanguino M, Zorrilla S, Keating CD, Monterroso B, Rivas G, Encapsulation of a compartmentalized cytoplasm mimic within a lipid membrane by microfluidics, Chem. Commun 53 (2017) 4775–4778. [DOI] [PubMed] [Google Scholar]
- [31].Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, Weis K, DEAD-box ATPases are global regulators of phase-separated organelles, Nature 573 (2019) 144–148. [DOI] [PubMed] [Google Scholar]
- [32].Cabre EJ, Monterroso B, Alfonso C, Sanchez-Gorostiaga A, Reija B, Jimenez M, Vicente M, Zorrilla S, Rivas G, The nucleoid occlusion SlmA protein accelerates the disassembly of the FtsZ protein polymers without affecting their GTPase activity, PLoS One 10 (2015), e0126434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Du S, Lutkenhaus J, SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD, PLoS Genet. 10 (2014), e1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gardner KA, Moore DA, Erickson HP, The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide, Mol. Microbiol 89 (2013) 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Buske PJ, Levin PA, A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo, Mol. Microbiol 89 (2013) 249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kroschwald S, Munder MC, Maharana S, Franzmann TM, Richter D, Ruer M, Hyman AA, Alberti S, Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery, Cell Rep. 23 (2018) 3327–3339. [DOI] [PubMed] [Google Scholar]
- [37].Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA, Stress-triggered phase separation is an adaptive, evolutionarily tuned response, Cell 168 (2017), 1028–1040 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Oltrogge LM, Chaijarasphong T, Chen AW, Bolin ER, Marqusee S, Savage DF, Multivalent interactions between CsoS2 and Rubisco mediate alpha-carboxysome formation, Nat. Struct. Mol. Biol 27 (2020) 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Snead WT, Gladfelter AS, The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation, Mol. Cell 76 (2019) 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Robles-Ramos MA, Margolin W, Sobrinos-Sanguino M, Alfonso C, Rivas G, Monterroso B, Zorrilla S, The nucleoid occlusion protein SlmA binds to lipid membranes, mBio 11 (2020) e02094–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Spitzer J, Poolman B, The role of biomacromolecular crowding, ionic strength, and physicochemical gradients in the complexities of life’s emergence, Microbiol. Mol. Biol. Rev 73 (2009) 371–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Baranova N, Radler P, Hernandez-Rocamora VM, Alfonso C, Lopez-Pelegrin M, Rivas G, Vollmer W, Loose M, Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins, Nat. Microbiol 5 (2020) 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J, GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis, Science 355 (2017) 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yu J, Liu Y, Yin H, Chang Z, Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters, Cell Discov 5 (2019) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Laskowska E, Kuczynska-Wisnik D, New insight into the mechanisms protecting bacteria during desiccation, Curr. Genet 66 (2020) 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kusuma KD, Payne M, Ung AT, Bottomley AL, Harry EJ, FtsZ as an antibacterial target: status and guidelines for progressing this avenue, ACS Infect Dis 5 (2019) 1279–1294. [DOI] [PubMed] [Google Scholar]
- [47].Fisher RA, Gollan B, Helaine S, Persistent bacterial infections and persister cells, Nat Rev Microbiol 15 (2017) 453–464. [DOI] [PubMed] [Google Scholar]
- [48].Monterroso B, Reija B, Jimenez M, Zorrilla S, Rivas G, Charged molecules modulate the volume exclusion effects exerted by Crowders on FtsZ polymerization, PLoS One 11 (2016), e0149060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gonźalez JM, Jiménez M, Vélez M, Mingorance J, Andreu JM, Vicente M, Rivas G, Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment, J. Biol. Chem 278 (2003) 37664–37671. [DOI] [PubMed] [Google Scholar]
- [50].Reija B, Monterroso B, Jimenez M, Vicente M, Rivas G, Zorrilla S, Development of a homogeneous fluorescence anisotropy assay to monitor and measure FtsZ assembly in solution, Anal. Biochem 418 (2011) 89–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding authors upon reasonable request.


