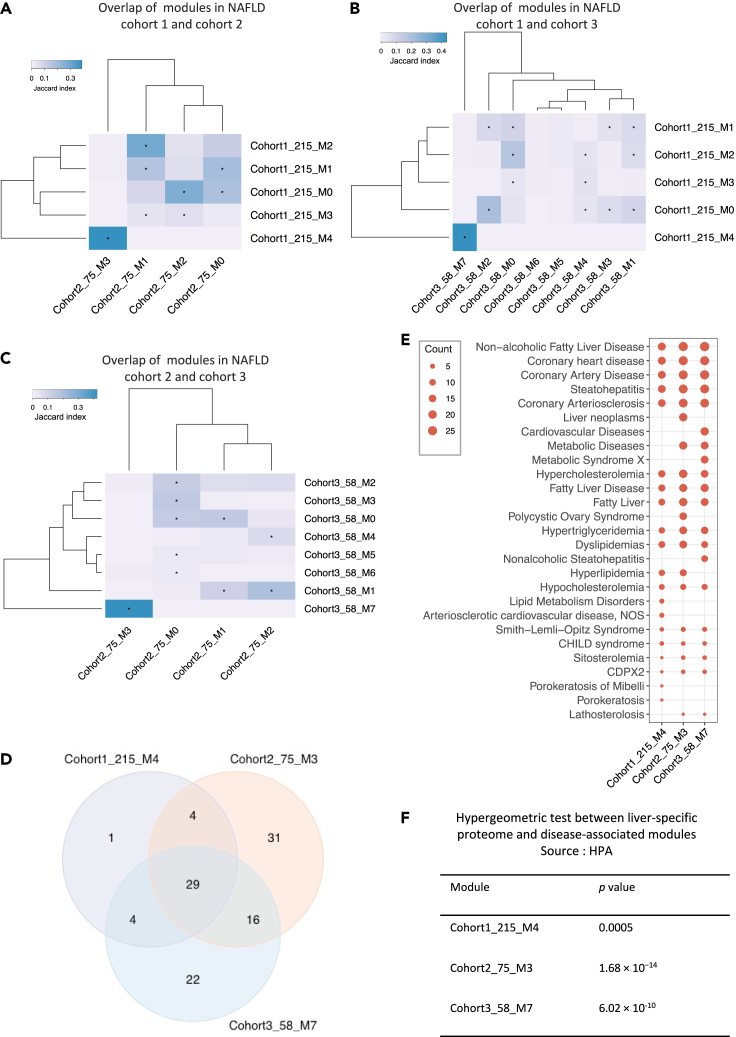
Figure 3.
Validation of disease-related modules using two independent NAFLD cohorts
(A–C) Hierarchical clustering of Jaccard Index between module pairs from NAFLD cohort 1 and 2; NAFLD cohort 1 and 3; NAFLD cohort 2 and 3. Color scales representing the range of the Jaccard index. Asterisk indicates the statistical significance of the overlap between gene members in any two modules from the different cohorts.
(D) Venn diagram shows numbers of genes overlapped between cohort1_215_M4, cohort2_75_M3, and cohort3_58_M7.
(E) Dot-plot heatmap shows top 20 significantly (“q-value FDR B&H” < 0.05) enriched diseases by genes in each module (cohort1_215_M4, cohort2_75_M3, and cohort3_58_M7). The size of each dot is proportional to the number of genes enriched in each disease term.
(F) The table shows the results from a hypergeometric test between liver-specific proteome (HPA) and disease-associated modules in NAFLD cohorts, the overlap with p value less than 0.05 was considered as significant.