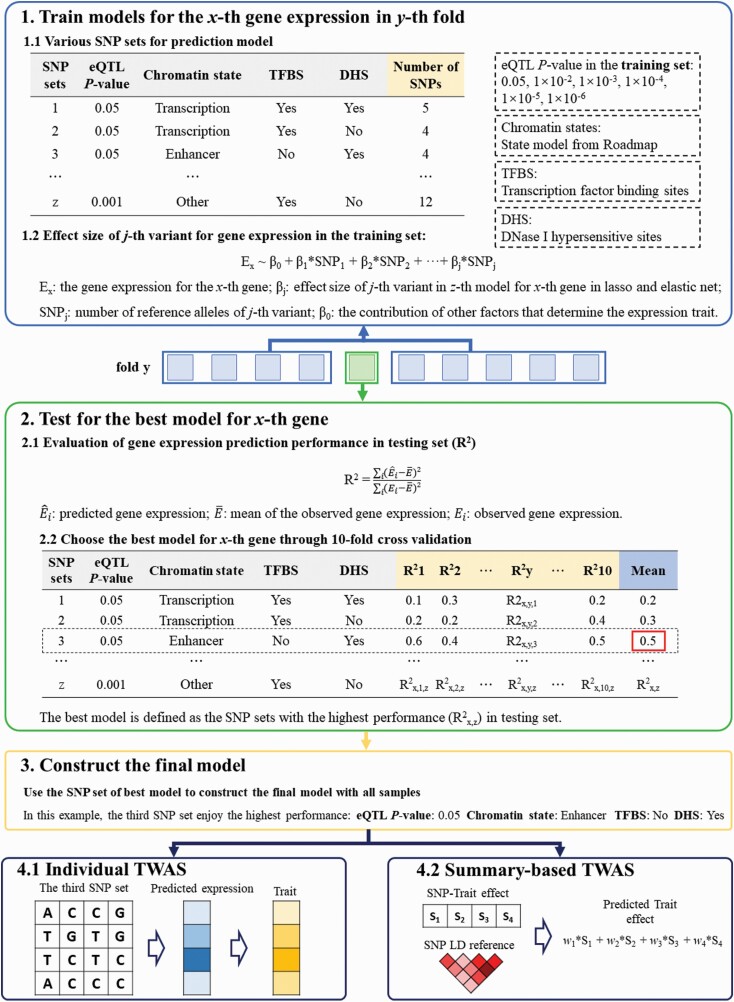
Fig. 1.
Schematic of ETWAS approach. The prediction framework includes 4 sections. First, for each gene, we divide the SNPs within 1 Mb of the transcription start/end site of the gene into multiple SNP sets according to the eQTL P-value and epigenetic annotations. We construct multiple elastic net and lasso models with the SNPs in each SNP set using 10-fold cross-validation R2 in the initial reference data. Second, we evaluate the prediction performance of each SNP set by cross-validation R2 between the predicted and observed expression, and the SNP set with the highest mean R2 is selected as the best model. Third, we construct the final prediction model with the parameters of the best SNP set using all the samples in the reference data. We then estimate the associations between predicted expressions and traits with the combination of SNP-trait effect sizes while accounting for linkage disequilibrium among SNPs.