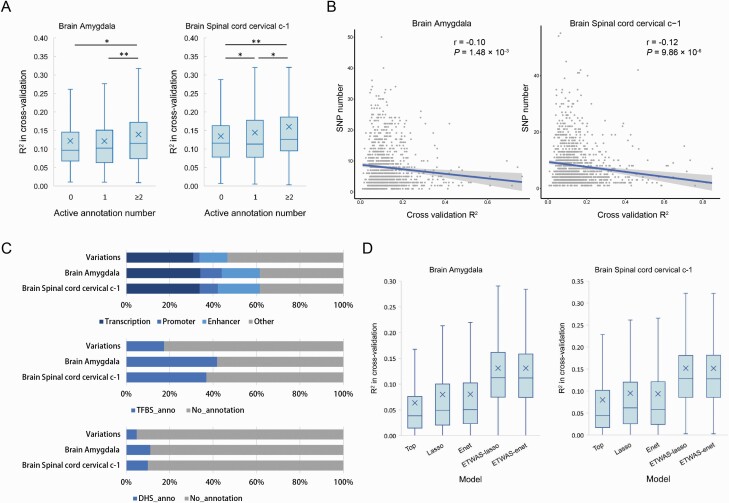
Fig. 2.
Epigenetic data improves the performance of gene expression prediction in brain tissues. (A) The cross-validation R2 of the best prediction models are sorted according to the active annotation number and group into 3 categories: 0, 1, ≥2. One asterisk (*) indicates P-value smaller than 0.05 (P < 0.05), 2 asterisks (**) indicates P-value smaller than 0.01 (P < 0.01). (B) The correlation between the prediction performance and the SNP number in the best model. The x-axis represents cross-validation R2 and the y-axis represents the SNP number of the best model. (C) The epigenetic annotation distributions of the SNPs used for all genes and the best ETWAS models in 2 brain tissues. (D) Accuracy of individual-level expression imputation models. Accuracy is estimated using cross-validation R2 between predicted and true expression. Box plots indicate the accuracy distribution for 2 brain tissues in 5 methods: top, lasso, elastic net (Enet), ETWAS-lasso, and ETWAS-enet.