Figure 4.
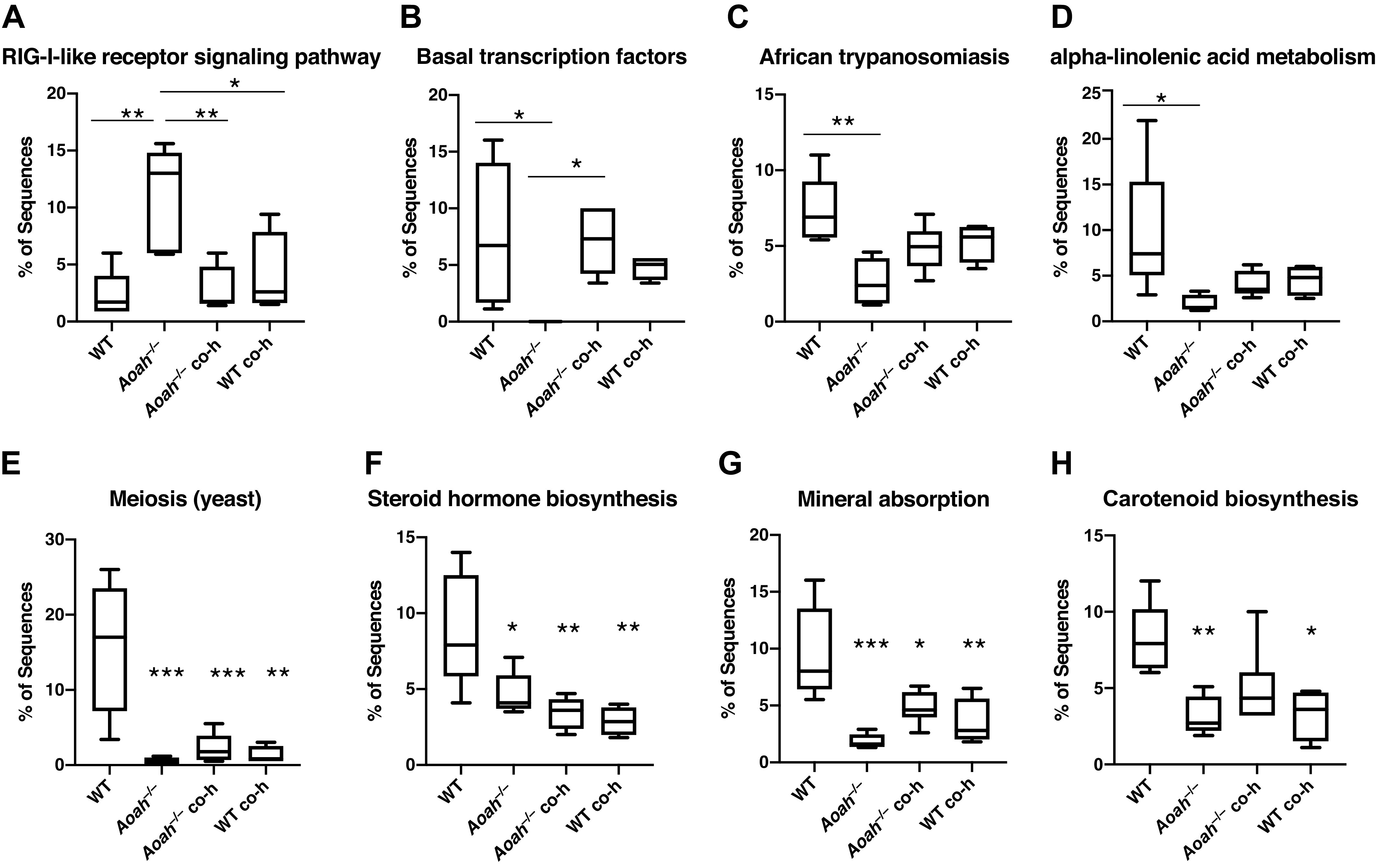
Microbiome-dependent changes in the functional characterization of bacterial communities. Biological functions of bacterial communities were predicted by implementing Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analyses using 16S rRNA sequence data from wild-type and AOAH-deficient fecal samples. Changes in the gut microbiome by genetic manipulation (Aoah−/−) or by cohousing (Aoah−/− co-h and B6 co-h) resulted in altered predictions including functions related to the retinoic acid-inducible gene I (RIG-I)-like receptor signaling pathway (A), basal transcription factors (B), African trypanosomiasis (C), α-linolenic acid metabolism (D), meiosis (yeast, E), steroid hormone biosynthesis (F), mineral absorption (G), and carotenoid biosynthesis (H). n = 5, *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA followed by post hoc Tukey’s HSD. Data represented as average ± SE. AOAH, acyloxyacyl hydrolase; co-h, cohoused; HSD, honestly significant difference.
