The oncogenic form of HSP90 organizes and maintains functional multienzymatic metabolic hubs in cancer cells, suggesting the potential of repurposing oncogenic HSP90 selective inhibitors to disrupt metabolism in lymphoma cells.
Abstract
HSP90 is critical for maintenance of the cellular proteostasis. In cancer cells, HSP90 also becomes a nucleating site for the stabilization of multiprotein complexes including signaling pathways and transcription complexes. Here we described the role of this HSP90 form, referred to as oncogenic HSP90, in the regulation of cytosolic metabolic pathways in proliferating B-cell lymphoma cells. Oncogenic HSP90 assisted in the organization of metabolic enzymes into non-membrane–bound functional compartments. Under experimental conditions that conserved cellular proteostasis, oncogenic HSP90 coordinated and sustained multiple metabolic pathways required for energy production and maintenance of cellular biomass as well as for secretion of extracellular metabolites. Conversely, inhibition of oncogenic HSP90, in absence of apparent client protein degradation, decreased the efficiency of MYC-driven metabolic reprogramming. This study reveals that oncogenic HSP90 supports metabolism in B-cell lymphoma cells and patients with diffuse large B-cell lymphoma, providing a novel mechanism of activity for HSP90 inhibitors.
Significance:
The oncogenic form of HSP90 organizes and maintains functional multienzymatic metabolic hubs in cancer cells, suggesting the potential of repurposing oncogenic HSP90 selective inhibitors to disrupt metabolism in lymphoma cells.
Introduction
Diffuse large B-cell lymphoma (DLBCL) and Burkitt lymphoma (BL) are highly proliferative diseases that exhibit, accordingly, an increased rate of catabolic glucose and glutamine metabolism (1). DLBCL and BL cells augment their aerobic glycolysis rates to produce biosynthetic intermediates for biomass accumulation and secondarily to supplement bioenergetic needs, because the majority of ATP is produced in the mitochondria (2). This and other metabolic traits of aggressive B-cell lymphomas are determined by the activation of molecular mediators including MYC and MTOR/PI3K, among others (3–5). Protein upregulation and expression of alternative enzyme isoforms are well established mechanisms that facilitate the metabolic reprogramming effect of oncogenic drivers (6). However, the role of enzymes compartmentalization in this process is less understood (7). Compartmenting metabolic complexes allows the regulation of macromolecular crowding, which modulates protein folding, aggregation, and diffusion (8, 9). Regulated protein crowding is beneficial to activate metabolic pathways that are not constitutively present or require protein upregulation to be established (10). This has been recognized in proliferating cells by the binding of the glycolytic and purine biosynthesis enzymes to the cytoskeleton (11–14). In addition to greater regulatory control, multienzymatic complexes that limit substrate diffusion represent an advantage to cells by increasing the solvation capacity within the cytosol and optimizing funneling of metabolites (8, 15).
Proliferating cells endure the bioenergetic challenge of maintaining cell homeostasis while facilitating biomass accumulation that enables cell growth and division. Metabolic demands are increased even further in cancer cells because of the stress imposed by an aberrant biological background and unstable microenvironmental conditions (1, 6). To increase the efficiency of the cellular metabolism, stressors promote the regulated crowding of metabolic proteins. For instance, under nutrient or hypoxic stress certain glycolytic enzymes coalesce into cytosolic clusters or “glycosomes” providing higher rates of glycolysis (16, 17). In cells cultured under purine metabolic stress, multiple enzymes catalyzing de novo purine biosynthesis colocalize to intracellular foci known as “purinosomes” to maintain the purines output (18–20). Moreover, IMPDH2 (inosine monophosphate dehydrogenase) of the purine biosynthesis pathway physically associates with CTPS1 (CTP synthase) of the pyrimidine biosynthesis pathway in cells grown under specific amino acid starvation (21). Although some enzymatic complexes can be self-regulated, others require the assistance of the stress chaperones HSP90 and HSP70 (16, 22, 23).
The “chaperome,” an assembly of molecular stress chaperones and their many partners, assist in protein folding and in reducing protein aggregation to maintain cellular proteostasis. In cancer cells, the “HSP90 chaperome” can be incorporated with the “HSP70 chaperome” to assemble higher-order structures regarded as “epichaperomes” (24). Epichaperomes can increase the fitness of the proteome of cancer cells by assisting in the formation of multiprotein complexes (24, 25). This function becomes critical for cancer proliferation on a background of internal and external stress that results from biological instability and changing microenvironmental conditions (1, 6). PU-H71 is an HSP90 inhibitor that kinetically selects for the HSP90-containing epichaperomes (i.e., oncogenic HSP90) over other HSP90 pools (24) and dismantles these multimeric long-lived complexes into individual, folding chaperones (26).
In this study, we described the role of oncogenic HSP90 (24) in the organization of metabolic enzymes into cytosolic nonmembrane-bound functional compartments in B-cell lymphoma cells. We demonstrated that this metabolic function of oncogenic HSP90 is required to fulfill biomass, energetic, and secretory requirements of B-cell lymphomas and, specifically, to support the metabolic program driven by MYC.
Patients and Methods
Human studies
Serum from de-identified healthy volunteers and patients with DLBCL were obtained with the approvals of the Institutional Review Boards (IRB) of Weill Cornell Medicine and Saint Louis University Hospital. Serum from healthy volunteers were selected to match the gender and age distribution of DLBCL patients in a 1:2 ratio. De-identified tonsillectomy specimens, BL and DLBCL tissues were obtained with the approvals of the IRB of Weill Cornell Medicine or University of Turin. Plasma from patients with DLBCL enrolled in the clinical trial NCT01393509 were obtained with the approval of the IRB of Memorial Sloan Kettering Cancer Center. Research conducted in accordance with Helsinki's Declaration. Patients provided written informed consent.
Mouse studies
All animal procedures were approved by the Research Animal Resource Center of WCM. OCI-Ly7 and Toledo DLBCL cells were injected in the flank of 24 severe combined immunodeficiency mice purchased from the US NCI. Once tumors reached 200 mm3, mice were treated with vehicle (0.9% sodium chloride solution, n = 6 for each cell line) or 75 mg/kg PU-H71 (n = 6 for each cell line) for 6 hours for protein proximity ligation assay (PLA) assays (OCI-Ly7 only) and 24 hours for metabolic assays. At the end of the experiment, mice were euthanized by CO2 inhalation and blood obtained post-mortem from cardiac puncture.
Chemical affinity purification of the oncogenic HSP90 interactome
PU-H71 and control (2-methoxyethlamine) beads were synthesized as reported previously (27). Before use, beads were washed three times in Felts buffer (20 mmol/L HEPES, 50 mmol/L KCl, 5 mmol/L MgCl2, 0.01% NP-40, freshly prepared 20 mmol/L Na2MoO4, and cOmplete protease inhibitors; Roche). One to 2 × 106 cells were lysed in 0.5 mL Felts buffer by rotation at 4°C for 30 minutes and vortexing for 10 seconds, followed by centrifugation at 14,000 × g for 5 minutes at 4°C. Supernatants were precleared with 100 μL of control beads for 1 hour at 4°C. After preclearing, half of each sample was incubated with 100 μL of either control or PU-H71 beads at 4°C overnight in the dark. Following incubation, bead conjugates were washed three times with Felts buffer, boiled in Laemmli buffer, and resolved by SDS-PAGE by standard immunoblotting procedure. The proteomics methodology was published in ref. 28.
Cell lines studies
Lymphoma cell lines OCI-Ly1 and OCI-Ly7 were cultured in Iscove's modified Eagle Medium (IMDM) with 20% FBS. Karpas422, Toledo, SU-DHL-6, and Raji were cultured in RPMI1640 with 10% FBS and 1% HEPES. These cell lines were obtained from the Ontario Cancer Institute, DMSZ, or ATCC and regularly tested for Mycoplasma sp. contamination by PCR. Annual cell identification was carried out by short tandem repeat analysis at the University of Arizona Genetics Core. P493–6 cells were obtained from the original developer Dr. Dirk Eick. Cells were maintained with 1% penicillin/streptomycin in a 37°C, 5% CO2, humidified incubator.
Cell number determination
All cell lines were grown at densities that allowed to maintain vehicle-treated cells in exponential growth over the drug exposure time. Relative cell number was determined by a fluorescence assay based on the measurement of a constitutive protease activity restricted to intact viable cells and that is independent of the cellular metabolic activity (CellTiter-Fluor; Promega). The fluorescence signal was kept within the linear range and was proportional to cell number. For the measurements we used a Synergy4 microplate reader (BioTek).
Immunoblotting
Cells were lysed in protease inhibitor-supplemented RIPA buffer by rotation at 4°C for 30 minutes, followed by centrifugation at 14,000 × g for 5 minutes at 4°C. Protein concentration was determined by BCA assay (Pierce Biotechnology) according to manufacturer's instructions. Protein lysates (10–20 μg) were resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with the indicated primary antibodies: α-tubulin (ab4074, Abcam), β-actin (AC-15, A3854, Sigma), BLNK (2B11, sc-8003, Santa Cruz Biotechnology), CAD (RB18384, AP11110c-ev, Abgent), CLPP (B-12, sc-271284, Santa Cruz Biotechnology), CTPS1 (C-13, sc-131474, Santa Cruz Biotechnology), G6PD (ab76598, Abcam), GAPDH (6C5, MAB374, Millipore Sigma), hexokinase 2 (C-14, sc-6521, Santa Cruz Biotechnology), HSP70 (W27, sc-24, Santa Cruz Biotechnology), HSP90 α/β (F-8, sc-13119, Santa Cruz Biotechnology), IMPDH2 (EPR8365B, ab129165, Abcam), and MTAP (EPR6893, ab126770, Abcam). Membranes were then incubated with peroxidase conjugated secondary antibodies (sc-2004 and sc-2005, Santa Cruz Biotechnology). We used ECL Western Blotting Substrate (Pierce Biotechnology) according to manufacturer's instructions and the blots were visualized by autoradiography. Quantitative densitometry analysis of Western blot bands was performed employing Image J version 10.2 (NIH).
Metabolomic assays
DLBCL cell lines and tissues: Low-passage OCI-Ly1 and OCI-Ly7 cell cultures were maintained at 106 cells/mL in fresh medium the night before adding either vehicle (PBS) or PU-H71 (0.5 μmol/L) to the culture for 6 hours. At endpoint, cells were harvested, washed once with cold PBS, and snap frozen in nitrogen. Snap frozen DLBCL human tissues were kept at −140°C until processed into lysates. Serum: blood (obtained by venipuncture in humans and post-mortem by cardiac puncture in mice) was immediately processed to isolate the serum fraction and kept at −80°C till the moment of processing for metabolomics.
Untargeted metabolomics was run by Metabolon Inc. Briefly, measurements were performed by ultra-high-performance liquid-phase chromatography and gas-chromatography separation, coupled with tandem mass spectrometry. For validation, targeted profiling of polar metabolites was performed at Weill Cornell Medicine Proteomics and Metabolomics Core Facility (WCM-PMCF) by ultra-high-performance liquid-phase chromatography coupled with tandem mass spectrometry followed by metabolite identification and quantitation.
Protein proximity ligation assay
P493–6 cells in absence of MYC and upon its induction by doxycycline withdrawal for 1, 3, and 6 hours, OCI-Ly1 cells treated with vehicle or 500 nmol/L PU-H71 for 3 and 6 hours, and lymphoma and bone marrow cells isolated from OCI-Ly1 xenografted mice treated with PU-H71 75 mg/kg for 6 hours were spun down at 250 × g for 5 minutes, plated on 8 mm coverslips previously coated with Cell-Tak, and incubated for 15 minutes at 37°C to adhere. After washing with PBS, they were fixed with 4% paraformaldehyde and permeabilized with PBS-Tween 0.5%. Cells were then subjected to the PLA assay using the Duolink Red Kit (O-link Bioscience, Sweden) in a humidity chamber according to the manufacturer's instructions. Briefly, cells were blocked with Duolink blocking buffer for 30 m and incubated for 1 hour at 37°C in the presence of primary antibodies for IMPDH (F-6, sc-166551, Santa Cruz Biotechnology), CTPS1 (sc-131474, Santa Cruz Biotechnology), GPI (HPA042620, Sigma-Aldrich), or RPIA (NBP2–02541, Novus Biologicals) diluted in Duolink antibody diluent at 1:100. After washing with Duolink washing buffer, cells were incubated with plus and minus PLA probes for 1 hour at 37°C followed by ligase addition and incubation for 30 minutes at 37°C. After washing, samples were then incubated in the presence of Duolink polymerase buffer containing the polymerase for 100 minutes at 37°C. Finally, covers were mounted with Fluoromount-G (Electron Microscopy Sciences) for laser confocal confocal microscopy. Cell images were acquired using a confocal microscope (Zeiss LSM880) with a Plan-Apochromat 63×/1.40 oil objective. To scan the whole cellular volume, 10 images at 0.2-μm step size were taken per field. Only clearly identifiable dots in the maximum intensity Z projection within the cell boundary were considered as indicators of protein proximity and the number of immobilized complexes per cell was determined.
Mass accumulation rate
We measured the mass accumulation rates (MAR) of individual cells using a serial suspended microchannel resonator (sSMR) device, as described previously (29, 30). OCI-Ly1 cells were cultured in IMDM with 20% FBS, then resuspended in their own medium at a concentration of 5 × 105 cells/mL for measurement. Cells were sampled from this untreated population and weighed 12 times over a 30-minute interval as they flowed through the sSMR device. After 1.5 hours, 1 μmol/L PU-H71 was added to the cell suspension and sampling of the population by the sSMR continued for ∼4 hours. A total of 106 cells were measured pretreatment and 161 cells posttreatment.
Substrate's consumption and lactate secretion
Low-passage cells were washed twice in PBS and resuspended at 106 cells/mL in RPMI without glucose or glutamine supplemented with 5.55 mmol/L glucose, 1 mmol/L glutamine, and 1% HEPES. A well without cells and with the same medium under the same culture conditions was used as reference to estimate the glucose and glutamine consumed and the lactate secreted in the wells with cells. For the measurements we centrifuged the cells at 250 × g, 4°C, for 5 minutes and immediately snap froze the supernatant to avoid degradation until analyzing it in a BioProfile Basic (Nova Biomedical) according to manufacturer's instructions.
Oxygen consumption rate and extracellular acidification rate
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using the Seahorse XF96 Extracellular Flux Analyzer (Agilent). Low-passage cells were seeded on Cell-Tak (BD Biosciences) precoated XF96 plates at 105 cells/well in 175 μL of sodium bicarbonate-free XF Base (Agilent) media freshly supplemented with 10 mmol/L glucose, 2 mmol/L L-glutamine, and 1 mmol/L pyruvate. Plates were spun at 40 g with breaks off and incubated in the XF incubator without CO2 for 25 minutes to ensure cell attachment. Measurements were taken before the addition of any inhibitor (basal respiration) and after the sequential injection of 1 μmol/L oligomycin (for ATP-linked respiration), 0.5 μmol/L FCCP (for maximal respiration), and 1 μmol/L rotenone/antimycin A (for non-mitochondrial oxygen consumption).
Medium oxygen consumption
Medium oxygen levels were measured in an SDR SensorDish Reader (PreSens). Cells were washed twice with PBS and changed to RPMI without glucose and glutamine (Biological Industries) and with 10% dialyzed FBS (Gibco) at 106 cells/mL. They were plated at 0.8 mL of culture/well on prewarmed 24-well OxoDish (PreSens) plates with integrated oxygen sensors. Medium oxygen levels were measured for 40 minutes to control for interwell variability. The measurements were then paused to add either vehicle or 0.5 μmol/L PU-H71 to the corresponding wells, and the oxygen of the medium was monitored for 1 more hour, at which point, the measurement was paused again to add either PBS or 2 mmol/L glutamine (Gibco) to the corresponding wells. Measurements went on for up to a total of 4 hours under regular incubation conditions (37°C, 5% CO2). The measurements after addition of substrate were normalized to 1 by dividing by the levels of each well before addition of substrate.
DNA and protein syntheses
Low-passage cell cultures were maintained at 106 cells/mL in fresh medium and either vehicle (PBS) or PU-H71 were added to the culture for 5 hours, after which, the thymidine analog EdU was added (10 μmol/L). After 1 hour, 106 cells were harvested and processed according to manufacturer's instructions. DAPI was added (1 μg/mL) to the final cell suspension and cells were analyzed after at least overnight incubation for best results. The DNA synthesis data are the mean fluorescence of the EdU incorporated in S-phase cells. For protein synthesis, cells were treated similarly with vehicle or PU-H71 (5.5 hours). Then they were changed to RPMI without methionine (Gibco) and supplemented with 50 μmol/L of the methionine analog HPG for 30 minutes. They were harvested and processed according to manufacturer's instructions. The protein synthesis data are the mean fluorescence of the HPG incorporated in the healthy population of cells. In both cases, data were collected on a MACSQuant flow cytometer (Miltenyi Biotech) and analyzed using FlowJo version 10.0.6 (TreeStar Inc.).
Real-time reverse transcriptase-qPCR
Total RNA was purified using TRIzol Reagent (Thermo Fisher Scientific) following manufacturer's instructions and resuspended in RNAse-free water. cDNA was synthesized using high-capacity RNA-to-cDNA Kit (Applied Biosystems). SYBR Green FastMix (Quanta BioSciences) was used for the PCRs. Primer sequences were designed to span exon-exon junctions in Primer-BLAST (NCBI).
Metabolic tracing
OCI-Ly1 cells were maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% FBS and 1% penicillin/streptomycin until a density of 106 cells/mL was reached. Cells were then washed once with PBS and incubated in glucose-depleted DMEM for 1 hour prior to starting the tracing. After washing once with PBS, cells were incubated for 30 minutes, 3 hours, or 6 hours in the presence of either DMSO (vehicle) or PU-H71 (1 μmol/L) in conditional DMEM containing 2.5 mmol/L of tracer U-13 C6-glucose plus 10% dialyzed FBS. After the incubation time, cells were washed once with PBS and centrifuged, and the pellets were saved at −80°C. Metabolites were extracted from cells using 80% methanol. Targeted LC/MS analyses were performed on a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) coupled to a Vanquish UPLC system (Thermo Fisher Scientific). The Q Exactive operated in polarity-switching mode. A Sequant ZIC-HILIC column (2.1 mm i.d. × 150 mm, Merck) was used for separation of metabolites. Flow rate was 150 μL/min. Buffers consisted of 100% acetonitrile for A, and 0.1% NH4OH/20 mmol/L CH3COONH4 in water for B. Gradient ran from 85% to 30% A in 20 minutes followed by a wash with 30% A and equilibration at 85% A. Metabolites were identified on the basis of exact mass within 5 ppm and standard retention times. Metabolites and their 13C isotopologues were identified on the basis of standard retention times and exact mass within 5 ppm. Relative quantitation was performed on the basis of metabolite peak area. The studies were conducted by the WCM Metabolomics and Proteomics Facility.
Isolation of human B-cell populations
Tonsils were minced, and mononuclear cells were isolated using Ficoll Histopaque density centrifugation. Naïve B cells were separated by positive selection using AutoMACS system (Miltenyi Biotec) after incubation with anti-IgD-FITC (BD Pharmingen) followed by anti-FITC microbeads (Miltenyi Biotec). Germinal center B cells (GCB) were separated by positive selection with anti-CD77 (AbD Serotec) followed by mouse anti-IgM, IgG1 isotype (BD Pharmingen) and anti-mouse-IgG1 microbeads (Miltenyi Biotec). Naive and germinal center B-cell purity (>90%) was determined by flow cytometry analysis of surface IgD (BD Biosciences, 555778), CD77 (Bio-Rad, MCA579) and CD38 (BD Biosciences, 340439). GCB were further verified by expression of A4GALT and BCL6 by RT-qPCR.
Statistical and bioinformatic analysis
Metabolomics dataset analyses
All metabolite concentrations were converted to log2 prior to statistical analysis. Identified metabolites with more than 20% missing values were excluded. The remaining missing values were imputed by drawing from a normal distribution with the same mean and SD as the nonmissing values from the respective metabolite. No named metabolites and exogenous metabolites were excluded from analysis. Each metabolite was annotated with (i) one of seven major biochemical super-pathways (“amino acid,” “peptide,” “lipid,” “energy,” “carbohydrate,” “nucleotide,” and “cofactors and vitamins”), and (ii) one subpathway. For each group of metabolites belonging to a pathway, mean z-scores were computed as a measure of average activity over all members of the pathway. Differences in metabolites between experimental conditions were assessed by standard two-sample t tests. For the overlaying of proteins and metabolites onto the Kyoto Encyclopedia of Genes and Genomes (KEGG) map of human metabolism we used iPath (31).
Transcriptional datasets analyses
For The Cancer Genome Atlas (TCGA) DLBCL set, level 3 raw expression counts of 48 patients were downloaded from TCGA. The WCM DLBCL cohort and cell lines RNA-seq data were aligned using STAR (v2.3) to human reference genome (version hg19/GRCh37), and raw counts were calculated with HT-seq. Normalized read counts as provided by DESeq2 were extracted from the three datasets for posterior analyses. Spearman correlations were calculated in R and plotted with ggplot2. Forty-four samples corresponding to BL and PMBL profiled on Affymetrix HG U133 Plus 2 arrays were downloaded from http://llmpp.nih.gov/BL/ and were used as deposited by the authors.
Proteomics dataset analyses
Data from the chemical precipitation of HSP90 cargoes from the cytoplasm of DLBCL cell lines OCI-Ly1 and OCI-Ly7 were obtained from previous experiments in our lab (28) and the union of the HSP90 interactome in the two cell lines was analyzed by KEGG pathway enrichment analysis in WebGestalt (32). The HSP90 cargo proteins under the category “metabolism” were further analyzed by STRING performed with a minimum required confidence interaction score of 0.4 on a scale from 0 to 1, and by determining the top 10 overrepresented KEGG pathways.
Degree of association between proteins and metabolites
To compare sets of significantly changed metabolites and the HSP90 metabolic interactome, metabolites and proteins were mapped to the Recon 2.04 metabolic network that contains metabolites annotated to genes in metabolic pathways. A metabolite and a protein were “connected” if their respective annotated reaction occurred in the same pathway.
Protein structural and biochemical properties analysis
We used ProtParam to compute the following protein properties: instability index, aliphatic index, and number of amino acids. We used UniProtKB to retrieve the adenosine nucleotide binding information, and we used PASTA2 to compile secondary structure predictions and intrinsic disorder.
Data availability
RNA-sequencing data for WCM patients with DLBCL are available at Gene Expression Omnibus (GEO) with the accession number: GSE145043. The metabolomics dataset of patients with DLBCL and healthy volunteers is available in Supplementary Materials and Methods.
Results
Oncogenic HSP90 nucleates multiprotein complexes containing metabolic enzymes
To study the function of oncogenic HSP90 in lymphoma cells, we took advantage of a method that we previously demonstrated as able to maintain the biochemical stability of HSP90-containing epichaperome and its associated interacting proteins (i.e., the HSP90 interactome; refs. 25, 33, 34) based on affinity purification of the native complex with a chemical bait followed by mass spectrometric identification of its components (HPLC-MS/MS; Fig. 1A; ref. 28). This methodology does not require exogenous protein expression while, unlike antibody-based precipitation, isolating only the oncogenic higher-order form of HSP90 multiprotein complexes (25). Pathway enrichment analysis of the oncogenic HSP90 interactome in the cytoplasm fraction of DLBCL cell lines OCI-Ly1 and OCI-Ly7 revealed a significant overrepresentation of proteins from cellular processes typically involving multiprotein complexes such as signaling, protein degradation, RNA translation and, strikingly, several cellular metabolic pathways (Fig. 1A). We and others have previously described the role of oncogenic HSP90 in maintaining the active conformation of the BCR signalosome, the BCL6 repressosome, and polyribosomes (28, 34, 35); however, the role of oncogenic HSP90 in the cellular metabolism of B-cell lymphomas has not been addressed.
Figure 1.
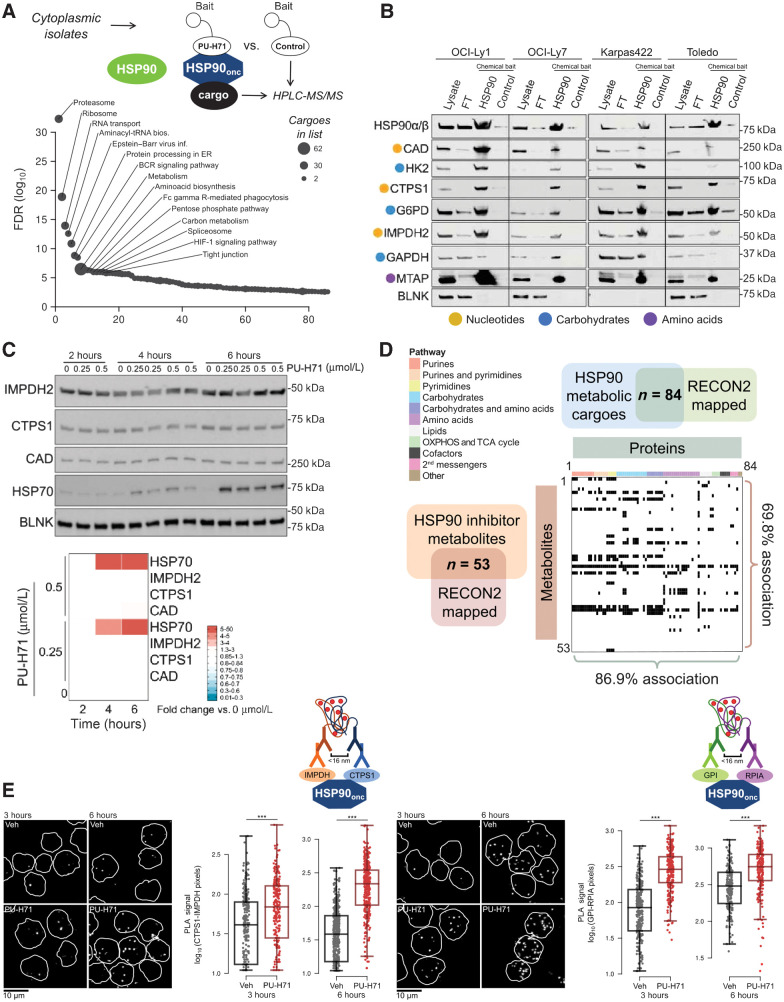
Functional association of the oncogenic HSP90 metabolic interactome and the metabolome in DLBCL. A, Pathways enriched in the oncogenic HSP90 (HSP90onc) interactome from the cytoplasmic fraction of OCI-Ly1 and OCI-Ly7 DLBCL cells. Proteins were identified by HPLC-MS/MS. Pathway enrichment as number of cargoes is indicated by dot size and their relative significance by the adjusted P value as FDR. B, Validation of representative enzymes from the oncogenic HSP90 interactome belonging to the three enriched metabolic pathways (indicated by colored dots) in the DLBCL cell lines OCI-Ly1, OCI-Ly7, Karpas 422, and Toledo. The lanes represent original cytosolic lysates, proteins remaining after oncogenic HSP90 chemical affinity purification (flow-through, FT), oncogenic HSP90 chemical affinity purification (HSP90), and inert chemical affinity purification (control). HSP90 was used as positive control and BLNK was used as non-HSP90-cargo negative control. C, Time course of the abundance of oncogenic HSP90 metabolic interactome components IMPDH2, CTPS1, and CAD upon oncogenic HSP90 inhibition with increasing concentrations of PU-H71 (0.25 and 0.5 μmol/L) or vehicle (0) in OCI-Ly1 cells. HSP70 was used as molecular readout of effective HSP90 inhibition and BLNK as a non-HSP90-cargo negative control. Densitometry of blots are shown at the bottom as color-coded fold changes over vehicle control. D, Association of the oncogenic HSP90 metabolic interactome mapped to RECON (n = 84) with the differentially expressed metabolites upon PU-H71 treatment mapped to RECON (n = 53). Proteins are ordered and color-coded by pathways. Matched protein-metabolite pairs are shown as black rectangles in the association plot. E, Representative images of IMPDH2-CTPS1 and GPI-RPIA endogenous complexes in OCI-Ly1 DLBCL cells exposed to vehicle (Veh) or PU-H71 for 3 and 6 hours. Bar, 10 μm. Quantifications of the PLA signal are shown next to images, with each dot belonging to a single-cell measurement. P values were calculated by t test. ***, P < 0.001.
To elucidate a potential role of oncogenic HSP90 in regulating the cellular metabolism, we first categorized its metabolic interactome into functional association networks. The metabolic interactome was significantly enriched for enzymes involved in the metabolism of nucleotides (purines and pyrimidines), carbohydrates, and amino acids (Supplementary Fig. S1A; Supplementary Table S1). We independently validated G6PD, GAPDH, and HK2 from “carbohydrates,” CAD, IMPDH2, and CTPS1 from “nucleotides” and MTAP from “amino acids” by chemical affinity purification followed by immunoblotting in OCI-Ly1, OCI-Ly7, Karpas422, and Toledo DLBCL cell lines (Fig. 1B). The cytosolic protein BLNK was used as a negative control (Fig. 1B). The oncogenic HSP90 metabolic interactome was not constituted from particularly thermally unstable proteins as determined by their biochemical properties including melting temperature (36) analysis (Supplementary Figs. S1B and S1C). Accordingly, PU-H71 at anti-neoplastic doses of 200 and 500 nmol/L for up to 6 hours did not result in decreased abundance of the oncogenic HSP90 metabolic interactome components CAD, IMPDH2, and CTPS1 (Fig. 1C). In contrast, between 14 and 24 hours of inhibitor treatment was necessary to observe the typical decrease of these cargo proteins (Supplementary Fig. S1D). However, upregulation of HSP70 demonstrated on-target activity of PU-H71 as early as 4 hours after administration. BLNK was used as negative control (Fig. 1C).
The analysis of the reactions catalyzed by the oncogenic HSP90 metabolic interactome indicated an enrichment on key enzymatic steps, defined as irreversible, committed, and/or rate-limiting reactions particularly of the “nucleotide” and “carbohydrates” pathways (Supplementary Fig. S2). To determine how inhibition of the oncogenic form of HSP90 will affect the cellular metabolism of B-cell lymphoma cells, we integrated the oncogenic HSP90 metabolic interactome with cellular metabolomics performed in the same DLBCL cell lines by bioinformatic analysis (37). Administration of 500 nmol/L PU-H71 for 6 hours (vs. vehicle control) to OCI-Ly1 and OCI-Ly7 cells caused a significant change in 90 metabolites (Supplementary Fig. S3A); and ∼70% of them were directly associated with at least one protein of the oncogenic HSP90 metabolic interactome (Fig. 1D), indicating a high degree of functional association. The full extent of the integrated proteomics and metabolomics data was visualized by overlaying it onto a canonical metabolism map (Supplementary Fig. S3B).
The high overlap between the oncogenic HSP90 interactome and its metabolome in absence of significant protein degradation suggested that oncogenic HSP90 regulates cellular metabolism by a higher-order proteome organization. These data suggested that oncogenic HSP90 is implicated in the formation and stabilization of dynamic protein complexes other than those involved in signaling and transcription regulation as we have previously demonstrated (28, 33, 34). We thus investigated the role of oncogenic HSP90 in the assembly of endogenous enzymatic complexes in cells by “freezing” the complexes using PU-H71 that specifically bound oncogenic HSP90 over other HSP90 pools (24) followed by visualization of interacting enzymes by PLA assays. We investigated two pairs of critical enzymes for proliferating cells: the enzymes IMPDH2 of the purine biosynthesis with CTPS1 of the pyrimidine biosynthesis; and GPI (phosphohexose isomerase), which catalyzes the isomerization of glucose-6-phosphate and fructose-6-phosphate in the glycolysis, with ribose 5-phosphate isomerase A (RPIA) that catalyzes the reversible conversion of ribose 5-phosphate and ribulose 5-phosphate in the pentose phosphate pathway (PPP). IMPDH2-CTPS1 and GPI-RPIA PLA complexes were analyzed in proliferating OCI-Ly1 DLBCL cells exposed to PU-H71 500 nmol/L for 3 and 6 hours. There was no significant change on protein expression levels during this period (Supplementary Fig. S3C). We found that PU-H71 stabilized IMPDH2-CTPS1 and GPI-RPIA complexes in OCI-Ly1 cells (Fig. 1E), indicating that oncogenic HSP90 facilitates the formation and stabilization of dynamic metabolic multienzymatic complexes.
Oncogenic HSP90 inhibition decreases nutrient utilization in DLBCL cells
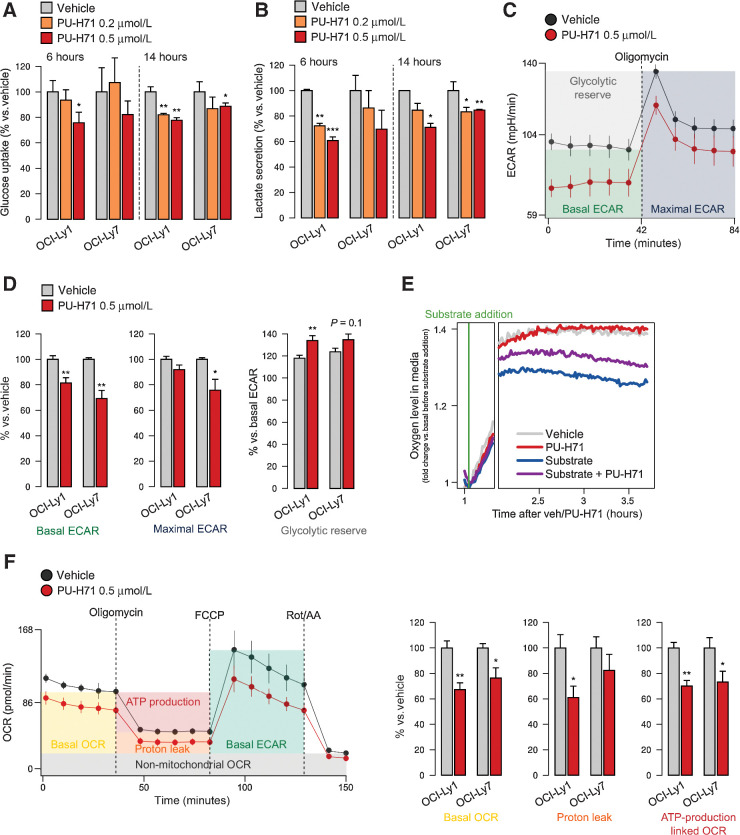
To determine the metabolic advantage provided by stabilization of dynamic multienzymatic complexes by oncogenic HSP90, we evaluated the effects of its inhibition on nutrient utilization at several levels. We measured the glucose uptake from the culture medium and the secretion of lactate as the end-product of glycolysis into the culture medium, in OCI-Ly1 and OCI-Ly7 cells treated with 200 and 500 nmol/L PU-H71 or vehicle for 6 and 14 hours. We found a decrease in glucose uptake (Fig. 2A) and lactate secretion (Fig. 2B) in both cell lines, compatible with impaired glycolysis. We independently confirmed this effect by measuring the ECAR in the same cell lines (Fig. 2C; Supplementary Figs. S4A and S4B). The basal ECAR was significantly lower in both cell lines (Fig. 2D). Upon addition of oligomycin to inhibit the mitochondrial respiration and elicit the maximal ECAR, cells treated with PU-H71 could not reach the maximal ECAR of vehicle-treated cells (Fig. 2D), leading to a significant increase in the glycolytic reserve by 15% to 20% (Fig. 2D). These data suggest that oncogenic HSP90 provides an increased efficacy to metabolize glucose in lymphoma cells.
Figure 2.
Oncogenic HSP90 inhibition decreases nutrient utilization. A and B, Glucose uptake (A) and lactate excretion (B) in OCI-Ly1 and OCI-Ly7 cells treated with vehicle, PU-H71 (0.2 μmol/L), and PU-H71 (0.5 μmol/L) for 6 and 14 hours. Data are normalized to vehicle-treated cells. C, ECAR of OCI-Ly1 cells treated with vehicle or PU-H71 at baseline and upon oligomycin treatment to estimate the maximal ECAR. Error bars, SD of 10 replicate wells. Representative experiment of triplicates is shown. D, Mean basal ECAR, maximal ECAR, and glycolytic reserve capacity in OCI-Ly1 and OCI-Ly7 cells treated with vehicle or PU-H71, normalized to vehicle. E, Mitochondrial respiration as determined by real-time measurement of oxygen levels in the tissue culture medium of OCI-Ly1 cells in absence of glutamine (substrate) with vehicle or PU-H71, and presence of glutamine with vehicle or PU-H71. Representative experiment of triplicates is shown. F, OCR of OCI-Ly1 cells treated with vehicle or PU-H71 at baseline upon oligomycin treatment to determine proton leak and OCR-linked ATP production, upon FCCP to estimate maximal OCR, and upon rotenone/antimycin-A (Rot/AA) to estimate non-mitochondrial OCR. Right, mean basal OCR, proton leak, and ATP production linked to OCR in OCI-Ly1 and OCI-Ly7 cells treated with vehicle or PU-H71. In all panels, unless stated differently, error bars are SEM of three independent experiments. P values were calculated by t test. n.s., not significant; *, P < 0.05; **, P < 0.01.
In addition to glucose, DLBCL cells consume glutamine in the mitochondria via glutaminolysis, a process that feeds the tricarboxylic acid cycle and readily increases mitochondrial respiration. We then measured in real-time the oxygen levels in the culture medium immediately after addition of glutamine as substrate in DLBCL cells treated with PU-H71. As expected, glutamine induced oxygen consumption (Fig. 2E), an effect reduced by PU-H71 (Fig. 2E). Similar results were obtained with the Burkitt lymphoma cell line Raji (Supplementary Fig. S4C). We independently confirmed the effects of PU-H71 on mitochondrial respiration by measuring the OCR in DLBCL cells cultured with glucose, glutamine, and pyruvate. We found that PU-H71 decreased basal OCR in OCI-Ly1 and OCI-Ly7 cells (Fig. 2F; Supplementary Figs. S4D and S4E), as well as respiratory parameters such as the proton leak and the ATP-production linked OCR in OCI-Ly1 cells (Fig. 2F). These data indicate that oncogenic HSP90 supports both glycolysis and mitochondrial respiration. The mechanism for this latter effect likely involves a more efficient channeling of substrates because PU-H71 does not penetrate mitochondria (38).
Oncogenic HSP90 inhibition decreases macromolecule biosynthesis and biomass gain
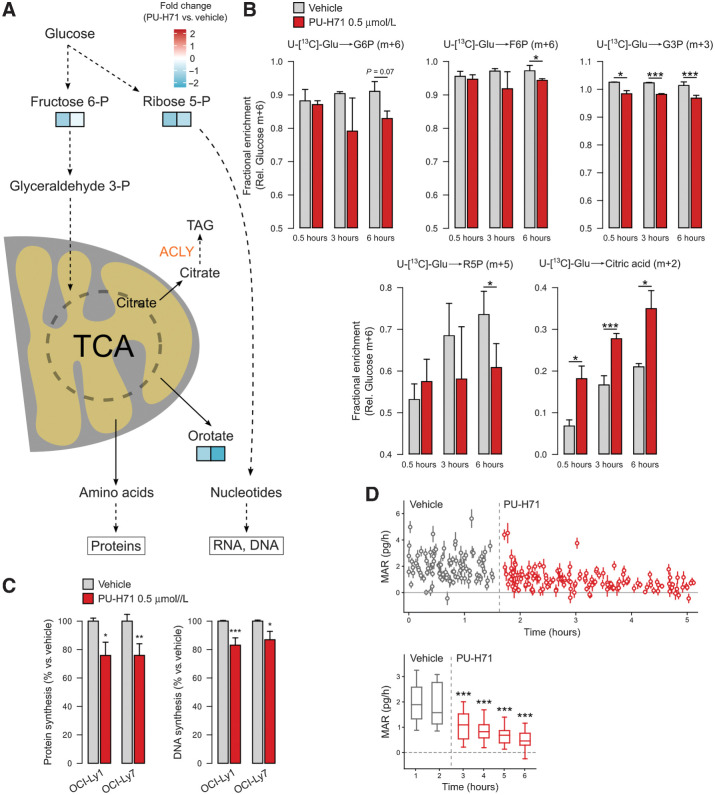
Although in non-proliferating cells respiration is mainly an ATP-producing catabolic process, in proliferating cells respiration serves also a crucial anabolic role by providing both direct and indirect intermediates, as well as cofactors, for various processes including synthesis of proteins and nucleotides (1). In this regard, the dihydroorotate dehydrogenase (DHODH) step of the pyrimidines biosynthesis is coupled to the mitochondrial electron transport chain and respiration. The de novo synthesis of nucleotides is also contingent on the ribose 5-phosphate formed primarily by the PPP. Pathway mapping of the metabolic changes caused by oncogenic HSP90 inhibition in OCI-Ly1 and OCI-Ly7 cells showed that key precursors fructose-6-phosphate, orotate and ribose 5-phosphate are decreased (Fig. 3A; Supplementary Figs. S5A and S5B). Furthermore, glucose carbon (D-glucose-13C6) tracing in OCI-Ly1 cells confirmed that glucose-derived glycolytic and PPP metabolites fructose-6-phosphate, ribose 5-phosphate, and glyceraldehyde-3 phosphate decreased upon HSP90 inhibition (PU-H71, 6 hour; Fig. 3B). Moreover, glucose-derived citrate increased in OCI-Ly1 cells treated with PU-H71 (Fig. 3B) potentially reflecting an attenuated activity of the key lipid biosynthesis enzyme ACLY, part of the oncogenic HSP90 metabolic interactome (Supplementary Figs. S1A and S1B). Alterations in the levels of precursors should be reflected in the biosynthesis of macromolecules. We thus measured the impact of oncogenic HSP90 inhibition on the synthesis of DNA and proteins by incubating cells with thymidine or methionine analogs, respectively, in presence of PU-H71 or vehicle. We found that inhibition of oncogenic HSP90 for 6 hours decreased DNA synthesis by 10% to 20% and protein synthesis by 25% in OCI-Ly1 and OCI-Ly7 cell lines (Fig. 3C).
Figure 3.
Oncogenic HSP90 inhibition decreases the synthesis of DNA, proteins, and biomass gain. A, Cartoon showing changes in selected metabolites from anabolic mitochondrial and pentose phosphate pathways and their metabolic fate in OCI-Ly1 and OCI-Ly7 cells. B, Glucose carbon tracing in OCI-Ly1 cells treated with vehicle or the oncogenic HSP90 inhibitor PU-H71 for 30 minutes, 3 hours, and 6 hours. The primary glucose-derived isotope for each metabolite is shown as relative to glucose m+6. The metabolite is indicated on top as glucose 6-phosphate (G6P), fructose 6-phosphate (F6P), glyceraldehyde 3-phosphate (G3P), ribose 5-phosphate (R5P), and citrate. C, Left, detection of newly synthesized protein by the incorporation rate of the amino acid analogue L-homoproparglyglycine in OCI-Ly1 and OCI-Ly7 cells treated with vehicle or PU-H71 0.5 μmol/L for 6 hours. Right, detection of newly synthesized DNA by the incorporation rate of thymidine analogue 5-ethynyl-2′-deoxyuridine in OCI-Ly1 and OCI-Ly7 cells treated with vehicle or PU-H71 0.5 μmol/L for 6 hours. In all panels, error bars are SEM of three independent experiments. D, Real-time assessment at single-cell resolution of cellular MAR in OCI-Ly1 cells treated with vehicle and upon administration of PU-H71 to the same culture. Bottom, mean MAR comparing binned datasets. P values were calculated by t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Altogether, these results suggested that lymphoma cells with an active form of oncogenic HSP90 can more efficiently utilize metabolites required for the synthesis of macromolecules, an effect that may facilitate the maintenance of the cellular biosynthetic activity under oncogenic stress. To noninvasively quantify the impact on the cellular biomass at single-cell resolution, we continuously monitored the mass accumulation rate (MAR; ref. 29) of OCI-Ly1 cells immediately after exposure to PU-H71 and for up to 6 hours. We found a significant reduction of MAR in cells with inhibited oncogenic HSP90. Strikingly, this effect occurred as early as 30 minutes after PU-H71 administration (Fig. 3D; Supplementary Fig. S5C), denoting a potential interference with nutrient utilization in DLBCL cells upon oncogenic HSP90 inhibition.
Oncogenic HSP90 supports the production of extracellular metabolites
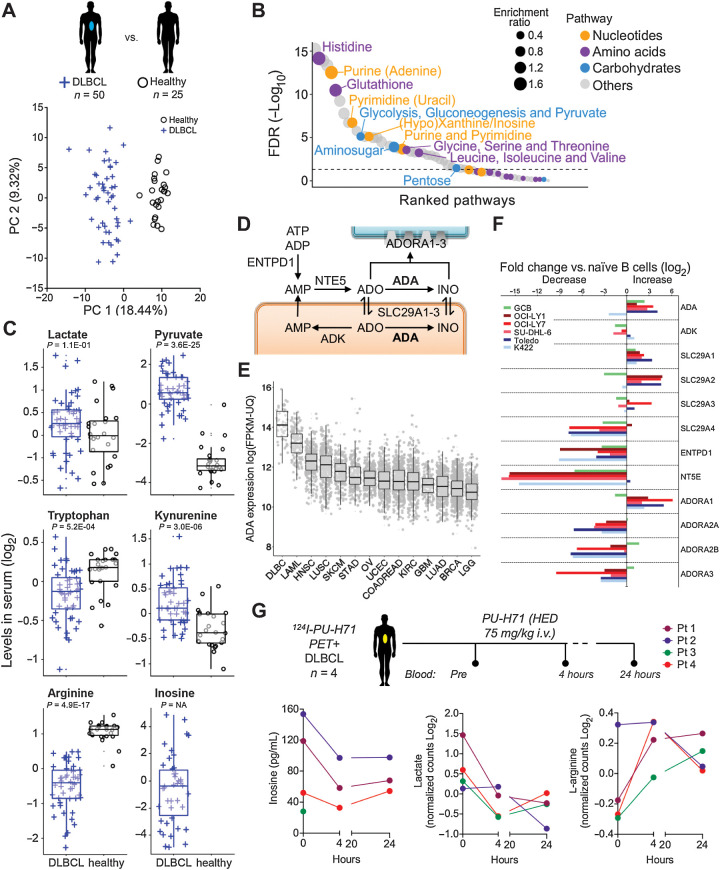
A further implication of our data in DLBCL cell lines is that, in addition to contributing to energy and biomass, certain oncogenic HSP90-dependent metabolites such as secreted lactate and purines could also have an impact on the tumor micro and macroenvironment. To contextualize this effect, we first determined the most active metabolic pathways in DLBCL by comparing the serum metabolomics of 50 DLBCL patients with 25 age- and gender-matched healthy individuals. The exometabolomics analysis readily segregated patients with DLBCL from healthy individuals (Fig. 4A). We identified 312 significantly different metabolites between these two groups (Fig. 4B; Supplementary Table S2), the majority belonging to the nucleotides, carbohydrates, and amino acids biochemical categories (Fig. 4B). A subgroup of these metabolites has been described, particularly in solid tumors, as regulating cancer immunity (39). We specifically analyzed these “immunometabolites” and found that patients with DLBCL had an exometabolome characterized by increased lactate and pyruvate, decreased tryptophan and increased kynurenine, decreased arginine and increased inosine (Fig. 4C). This pattern is compatible with an environment that depresses lymphoma immunity. Inosine represented the most extreme example of these metabolites as it was detected exclusively in patients with DLBCL (Fig. 4C). Extracellular inosine depends on adenosine levels, which in turn are contingent on the transport from the intracellular space by nucleoside transporters. Adenosine is deaminated to inosine by adenosine deaminase (ADA) (Fig. 4D), and both purines exert similar immune regulatory effects (40). Extracellular inosine is much more stable than adenosine, which goes in agreement with the lack of detection of adenosine in the serum from our cohorts of healthy or DLBCL individuals (Supplementary Table S2). The prevalence of inosine over adenosine in patients with DLBCL could be associated to the higher expression of ADA in comparison with other tumor types (Fig. 4E). To further characterize the inosine pathway in DLBCL, we determined the expression of the enzymes, transporters, and receptors related to inosine metabolism (Fig. 4D) in OCI-Ly1, OCI-Ly7, SU-DHL-6, Karpas422, and Toledo DLBCL cell lines and found a higher expression of ADA compared with normal human naïve B cells in most cell lines (Fig. 4F). Moreover, in three of the DLBCL cell lines, the expression was even higher than in highly proliferative normal CD77+ GCBs (Fig. 4F), suggesting that DLBCL cells may have increased capacity of producing inosine. More importantly, lymphoma cell lines overexpressed the adenosine-inosine equilibrating membrane transporters SLC29A1 and SLC29A2 (Fig. 4F), responsible for the presence of these purines in the extracellular space. The purinergic receptor ADORA1 was also increased in the lymphoma cell lines (Fig. 4F).
Figure 4.
The serum metabolomics of patients with DLBCL reflects a macro- and microenvironment characterized by the presence of purines. A, Principal component analysis of serum exometabolomics of patients with DLBCL (n = 50, blue cross) and age- and gender-matched healthy individuals (n = 25, black circles). B, Metabolic pathways significantly different between patients with DLBLC and healthy individuals. The relative size of the circles indicates enrichment ratio versus healthy. The color coding indicates super-pathway categorization into “amino acids,” “carbohydrates,” “nucleotides,” and “others.” C, Boxplots of immune-related metabolites from the serum exometabolomics analysis from A. Levels are shown normalized as log2. Adjusted P values for the comparison are shown below each metabolite. Inosine was not detected in healthy individuals. D, Inosine pathway indicating metabolites and intra- and extracellular enzymes. E, Expression of ADA in DLBCL versus other tumors included in the TCGA cohort. F, Expression of enzymes and solute transporters involved in the metabolism of inosine in OCI-Ly1, OCI-Ly7, SU-DHL-6, Karpas422, and Toledo DLBCL cell lines and normal GCB normalized to levels in naïve B-cells. Representative experiment shown. G, Levels of inosine, lactate, and arginine in four patients with DLBCL expressing the oncogenic form of HSP90 (as determined by 124I-PU-H71 PET-CT), before and 4 and 24 hours after the administration of one dose of PU-H71 at human-equivalent dose of 75 mg/kg in mice.
Similar to the effect described in vitro, administration of PU-H71 75 mg/kg (vs. vehicle) for 6 hours in mice implanted with OCI-Ly7 (n = 10) was associated with stabilization of IMPDH2-CTPS1 complexes in lymphoma cells (Supplementary Fig. S6A). There was no effect in normal hematopoietic cells obtained from the bone marrow in these mice, in agreement with a preferential effect of PU-H71 on the oncogenic form of HSP90 (34). To determine the effect of inhibiting oncogenic HSP90 in the secretion of immunometabolites, we quantified the serum levels of secreted adenosine and inosine in mice implanted with OCI-Ly7 (n = 12) or Toledo (n = 12) cells and, once tumors reached 200 mm3, treated with one dose of PU-H71 75 mg/kg or vehicle for 24 hours. We observed that adenosine and inosine decreased upon oncogenic HSP90 inhibition in both DLBCL models (Supplementary Fig. S6B). We further evaluated this effect by measuring inosine, lactate, and arginine in the plasma of 4 patients with DLBCL with tumors with high 124I-PU-H71 uptake, a feature that noninvasively indicates the presence of active oncogenic HSP90 in cancer tissues (25, 41, 42). Patients with DLBCL were intravenously administered a single dose of PU-H71 (equivalent to 75 mg/kg in mice), and plasma was obtained before and 4 and 24 hours after (26). We found that, for each metabolite, 3 of 4 patients had either a decrease in inosine and/or lactate and/or increase in L-arginine as soon as 4 hours after PU-H71 (Fig. 4G). In most cases the changes plateau at the 24 hours' time point (Fig. 4G). These findings indicate that oncogenic HSP90 contributes to sustaining the secretory metabolic needs of DLBCL.
Oncogenic HSP90 supports the metabolic program of MYC in lymphomas
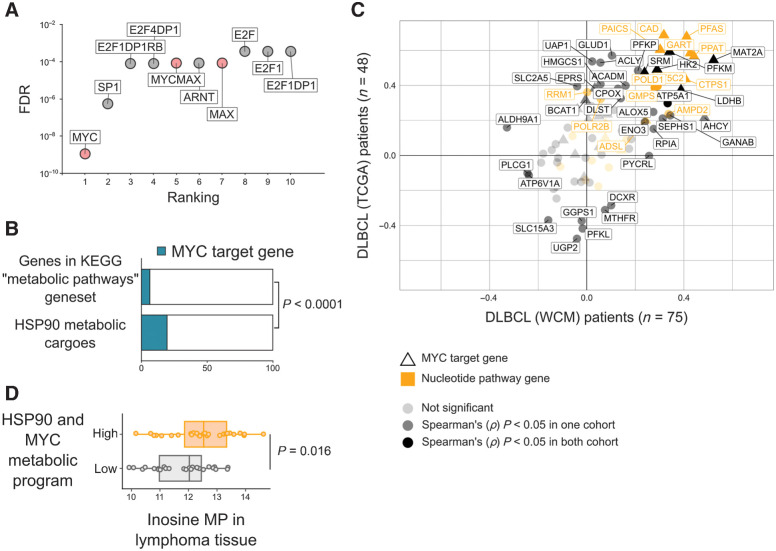
To understand the mechanistic relevance of these findings for the reprogramming of lymphoma metabolism, we analyzed putative transcription factors regulating the expression of the oncogenic HSP90 metabolic interactome. We found that the promoter of genes coding for the oncogenic HSP90 metabolic interactome were significantly enriched for consensus binding sites of MYC and MYC/MAX, SP1, E2F family, and ARNT (aryl hydrocarbon receptor nuclear translocator) (Fig. 5A), suggesting that these transcription factors may be more reliant on oncogenic HSP90 to reprogram cell metabolism. MYC is known to carry out a transcriptional program that includes numerous metabolic enzymes (43) that were significantly overrepresented in the oncogenic HSP90 metabolic interactome (Fisher exact P < 0.0001, Fig. 5B).
Figure 5.
The oncogenic HSP90 metabolic interactome supports the MYC metabolic program in DLBCL. A, Ranking of transcription factors regulating the oncogenic HSP90 metabolic interactome in DLBCL according to the presence of canonical binding sites on their promoter regions. Statistical significance was established by FDR. B, Oncogenic HSP90 metabolic interactome components presenting MYC binding sites in promoters compared with the universe of metabolic genes from the KEGG database containing MYC binding sites. Statistical enrichment was established by Fisher exact test. C, Spearman rank correlation (ρ) plots comparing the expression of MYC versus the expression of genes from the oncogenic HSP90 metabolic interactome. Correlations were conducted in two independent DLBCL patients’ cohorts composed of 48 TCGA cases (y-axis) and 75 WCM cases (x-axis). Canonical MYC target genes are depicted with triangles. Genes from the nucleotide's pathway are shown in yellow. Darker shading indicates significant (P < 0.05) correlations found in both cohorts. D, Whole tissue levels of inosine 5′-monophosphate (by HPLC-MS) in 54 DLBCL biopsies classified as high (n = 27) vs. low (n = 27) expression of the MYC metabolic program supported by oncogenic HSP90 determined by gene expression (RNA sequencing) correlation as in C.
MYC is an important prognosis-associated oncogene in DLBCL (1, 44, 45), which regulates a myriad of cellular pathways including metabolism. To determine the relevance of oncogenic HSP90 for the metabolic program of MYC, we correlated the expression of MYC and oncogenic HSP90 metabolic interactome genes in two independent RNA-sequencing datasets of 75 and 48 patients with DLBCL. We found 16 oncogenic HSP90 metabolic interactome genes that correlated significantly (P < 0.05) with MYC expression in at least one dataset (14/16 correlated in both datasets), with 57% of them from the nucleotides pathway and most of them representing direct MYC target genes (Fig. 5C; Supplementary Table S3). Strikingly, this correlation included practically all the enzymes required for the de novo synthesis of inosine such as PPAT, GART, PFAS, and PAICS (Fig. 5C; Supplementary Fig. S2; Supplementary Table S3). We thus determined the tissue levels of inosine 5-monophosphate in 54 DLBCLs by HPLC-MS. We found that DLBCLs expressing higher levels of the 16 correlated genes presented also significantly higher levels of inosine 5-monophosphate in these tissues (P = 0.016, Fig. 5D), indicating a functional association of the programs.
Recently, a transcriptomic-derived signature identified a group of MYC-driven DLBCLs termed DHITsig positive (45). DHITsig positive lymphomas are enriched for MYC and E2F target genes including metabolic genes as well as lower infiltration of CD4+ T cells (45). In our cohort, we found that in addition to lower CD4+ T cells, the microenvironment of these lymphomas has lower infiltration of CD8+ T cells and exhibits low immune cytolytic activity (Supplementary Figs. S7A and S7B; ref. 46). Taken together, this suggests that interference with the MYC metabolic program could not only impact cancer cells but also potentially improve lymphoma immunity.
MYC activation favors the assembly of oncogenic HSP90 complexes
MYC translocations resulting in high levels of MYC expression are considered a primary genetic event in BL (44). We thus investigated whether the expression correlation of MYC with the subset of transcripts corresponding to the oncogenic HSP90 metabolic interactome was also present in patients with BL. We compared the gene expression profiles of 24 patients with BL with 20 patients with primary mediastinal B-cell lymphoma (PMBL), a B-cell lymphoma not driven by MYC (44). Similar to DLBCL, we found a strong association between the expression of MYC, HSP90, (HSP90AB1) and the oncogenic HSP90 metabolic interactome in patients with BL, but not in patients with PMBL (Fig. 6A). Moreover, the nucleotide metabolism enzymes and direct MYC target genes IMPDH2, CTPS1, and CAD were among the most strongly positively correlated with MYC expression in patients with BL (Fig. 6A). We then analyzed if they were part of the oncogenic HSP90 interactome in BL by using PU-H71 bait in sequential purification experiments using primary lymphoma cells obtained from a patient with MYC-translocated BL (Fig. 6B). CLPP (47), a mitochondrial protein and MYC target gene, was used as negative control. We found that, like DLBCL cell lines, in primary BL cells the oncogenic form of HSP90 nucleated most of the IMPDH2, CTPS1, and CAD present in the sample (Fig. 6B). There was no binding of oncogenic HSP90 to CLPP, which reinforces our early observation that PU-H71 is not targeting the mitochondria directly.
Figure 6.
MYC activation in lymphoma cells induces the oncogenic HSP90 conformation to sustain metabolic complexes. A, Transcript expression heatmap of MYC, HSP90 (HSP90AB1), and HSP90 metabolic interactome components in MYC-dependent BLs and MYC-independent PMBLs. Canonical MYC target genes are marked with black rectangles and genes from the nucleotide's pathway with yellow rectangles. B, Majority of oncogenic HSP90 metabolic interactome components IMPDH2, CTPS1, and CAD are bound to oncogenic HSP90 in a BL patient sample presenting MYC translocation. HSP90 was used as positive control and CLPP as negative control. Chemically inert beads were used as control beads. C, Abundance of the oncogenic HSP90 conformation (higher-order interaction of HSP90 and HSP70 through HOP) in P493–6 BL cells according to MYC induction. Total protein abundance in cells (lysate) and the fraction corresponding to the oncogenic HSP90 conformation purified with PU-H71-beads in P493–6 cells in low MYC and upon its induction by doxycycline (Dc) withdrawal (6 hours). The quantification at the bottom indicates the abundance of the HSP70- and HOP-containing epichaperome complexes (denoted by a green square) normalized to the MYC-low (Dc+) state. D, Representative imaging of IMPDH2-CTPS1 (top) and GPI-RPIA (bottom) endogenous complexes in P493–6 BL cells in lower MYC (0 hours, Dc+) and higher MYC conditions (1, 3, and 6 hours after Dc withdrawal). Bar, 10 μm. Quantification of the IMPDH2-CTPS1 and GPI-RPIA endogenous complexes in P493–6 BL cells in the conditions described. n.s., not significant; ***, P < 0.001.
The oncogenic form of HSP90 is characterized by the enhanced physical integration of the HSP90 and HSP70 chaperomes into stable assemblies (i.e., epichaperomes) and emerges from changes in the cellular proteome driven by oncogene activation (25). To study how the establishment of the oncogenic HSP90 conformation may depend on MYC expression, we used B-cell lymphoma cells P493–6 carrying a conditional MYC allele. The epichaperome abundance was determined in MYC present/absent conditions by co-purification of oncogenic HSP90 with HSP70 (HSC70) and the adaptor protein HOP (HSC70/HSP90-organizing protein; ref. 25). P493–6 cells were treated with doxycycline (Dc) to downregulate MYC for 6 hours followed by 1 hour doxycycline withdrawal. In these conditions, there were no significant changes in cell proliferation and/or apoptosis between MYC− and MYC+ cells (Supplementary Fig. S7C). Although the total abundance of HSP90, HSP70, and HOP did not change significantly between MYC+ and MYC− cells, the epichaperome abundance increased in MYC+ cells (Fig. 6C), suggesting that MYC activity rapidly induces oncogenic HSP90 complexes. This translated into changes in the assembly of cytosolic multienzymatic metabolic complexes as shown by a significant increase in the number of IMPDH2-CTPS1 and GPI-RPIA complexes in MYC+ cells for up to 6 hours (Fig. 6D). There was no significant change on protein expression levels during this period (Supplementary Fig. S3C). These data indicated that establishment of a MYC-driven metabolism is favored by the presence of oncogenic HSP90. Taken together, our data demonstrated that inhibition of oncogenic HSP90 leads to disassembly of multienzymatic complexes ultimately causing the metabolic collapse of lymphoma cells providing an additional mechanism for the antineoplastic effect these inhibitors.
Discussion
In DLBCL and BL cells, HSP90 organizes into higher order complexes termed epichaperomes that are more stable than the classical folding chaperome complexes characteristic of normal cells (25). Contrary to classical transient chaperomes, whose function is to fold and stabilize proteins, the long-lived oncogenic HSP90 conformation allows maintenance of diverse protein complexes in active configurations (24, 25). We report here that the oncogenic form of HSP90 serves as a nucleating site for multienzymatic complexes in DLBCL and BL. By limiting the cytosolic distribution of functionally related enzymes, these assemblies or “metabosomes” increase the efficiency of metabolic pathways. We demonstrate that DLBCL cells with this oncogenic form of HSP90 possess a metabolic advantage to maintain their biomass and nutrient utilization. Although MYC and other transcription factors exert their oncogenic activity by increasing the expression of their target genes, there are several limitations to the extent to which a protein can be expressed without inducing detrimental molecular crowding (48). The nucleating capacity of oncogenic HSP90 adds to the regulation of protein crowding by favoring “metabosomes,” thus maximizing the activity of overexpressed enzymes.
Microcompartmentation, either due to membrane-restriction or by clustering of enzymes into functional complexes, has implications for efficiency of metabolic pathways and to segregate “moonlighting” functions of many enzymes and metabolites (49). In addition to the generation of gradients, the spatial organization of enzymes into complexes has been proposed to facilitate substrate funneling (8). Metabolism is thus accomplished through the spatial compartmentation of metabolic complexes into vesicles and organelles as occurs, for example, during fatty-acid oxidation and the tricarboxylic acid cycle (8). However, the demonstration of the functional metabolic microcompartmentation of the cytosol in mammalian cells has been more elusive. Cytosolic fluidity is subject to the dense lattice constituted by the cytoskeleton (48) and, accordingly, pharmacological disruption of actin and/or tubulin polymerization affects the cytosolic distribution of the cellular proteome (14, 50). However, the dynamic mechanism of organizing cytosolic multienzymatic complexes into functional microcompartments, rather than stochastic microcompartments as the mere distribution model imposed by the cytoskeleton would suggest, has remained incompletely understood. Taking advantage of our methodology to isolate native proteins nucleated by oncogenic HSP90, we characterized a network of functionally related metabolic enzymes from the cytosol of lymphoma cells. Similar to other chaperone-containing phase-separated granules (16, 49, 51, 52), it is possible that the transient, enzyme–enzyme interactions during catalysis become more stable in the context of oncogenic forms of HSP90 (24, 25, 49).
There is renewed interest in targeting aberrant tumor cell metabolism for cancer therapeutics. However, apart from selective compounds targeting mutant enzymes, drugs that can safely target metabolic pathways in tumors, as opposed to normal cells, have not been clearly identified (1, 53). Our study supports the notion of targeting not a single protein but rather the organization of the metabolic proteome to effectively decrease tumor metabolism (54). Overall decrease in tumor metabolism with HSP90 inhibitors specific for its oncogenic form is achieved earlier and at lower doses than those required to elicit protein degradation and apoptosis (34), a hallmark of all HSP90 inhibitors. This will offer the advantage of lower toxicity to normal tissues since normal cells do not contain epichaperomes (24, 25), and could also decrease the emergence of cell-intrinsic compensatory pathways. Despite the selectivity of PU-H71 for HSP90-containing epichaperomes, it is possible that the metabolic changes described here reflect both direct and indirect effects of HSP90 inhibition and further work is needed to confirm that all the observed changes are due to the specific inhibition of HSP90.
Metabolic reprogramming of cancer cells could, in turn, improve the anti-neoplastic activity of other therapeutic strategies. In this regard, our study shows that HSP90 activity in DLBCLs favors an inosine-rich environment. Inosine is an endogenous nucleoside produced by deamination of adenosine by ADA. Although inosine has a prolonged half-life in comparison with adenosine, neither nucleoside is usually detected in the plasma of healthy individuals. Through the direct activation of purinergic receptors in immune cells, inosine exerts immunomodulatory effects similarly to adenosine (55). Therefore, our results propose that oncogenic HSP90 inhibitors could deplete the lymphoma environment from the immunosuppressive effects of inosine and thus potentially improve lymphoma immunity and immunotherapy.
We also showed that the regulation of cellular metabolism through this mechanism is particularly relevant for lymphomas with MYC activation. In cancer cells, MYC enhances both catabolism and anabolism to meet the challenges of tumor growth (43). Taken together, our data support the overreaching concept that the oncogenic form of HSP90 optimizes several MYC metabolic pathways including the production of nucleotides. Other MYC activated pathways such as fatty acids and cholesterol metabolism seem to be secondarily dependent on the nucleating properties of oncogenic HSP90. This notion is supported by the relatively few, mostly cytosolic, lipid metabolic enzymes in the oncogenic HSP90 interactome compared with the extent of the alterations in the lipid metabolism as shown in the metabolomics experiments. Given the importance of MYC in driving metabolic reprogramming in other cancer types, our results bear promises as an alternative manner to target the aberrant metabolism characteristic of cancer cells.
Authors' Disclosures
E. Tikhonova reports personal fees from BostonGene LLC during the conduct of the study and personal fees from BostonGene LLC outside the submitted work. N. Kotlov reports personal fees from BostonGene LLC during the conduct of the study and personal fees from BostonGene LLC outside the submitted work; also has a patent for B1462.70024US00 pending and licensed to BostonGene. T. Taldone reports personal fees from Samus Therapeutics outside the submitted work. J.P. Leonard reports personal fees from Sutro, Bayer, Gilead/Kite, AstraZeneca, Celgene/BMS, ADC Therapeutics, Sandoz, Karyopharm, Miltenyi, Epizyme, Regeneron, GenMab, Abbvie, Incyte, Janssen, Eisai, and Mustang Bio; grants and personal fees from Roche/Genentech outside the submitted work. P. Martin reports personal fees from ADCT, AstraZeneca, Beigene, BMS, Gilead, Incyte, Janssen, Karyopharm, Morphosys, and Regeneron outside the submitted work. G. Chiosis reports nonfinancial support from Samus Therapeutics outside the submitted work; also has a patent for U.S. Patent No. 7,834,181 issued, licensed, and with royalties paid from Samus Therapeutics. S.R. Manalis reports personal fees from Travera and nonfinancial support from Affinity Biosensors outside the submitted work; also has a patent for US Patent 8,087,284 issued to Affinity Biosensors and Travera and a patent for US Patent 9,134,295 issued to Travera; and is a founder of Travera and Affinity Biosensors. L. Cerchietti reports grants from NCI, Doris Duke Charitable Foundation, and Leukemia and Lymphoma Society during the conduct of the study; grants from BMS/Celgene outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figures
Supplementary Table 1, Table 2 and Table 3
Acknowledgments
The authors thank Dr. Dirk Eick from the GSF-Research Center for Environment and Health, Munich, Germany, for providing P493-6 cells and Drs. Justin Cross and Vladimir Yong from the Marron Cancer Metabolism Center of Memorial Sloan Kettering Cancer Center for help with cellular OCR and ECAR measurements. The work involving human specimens was supported by the Doris Duke Charitable Foundation DDCF 2012070 (to L. Cerchietti). Additional work was supported by the Leukemia & Lymphoma Society SCOR award (LLS-SCOR 7012-16) component core C (to L. Cerchietti), the NIH 1U54CA217377 (to S.R. Manalis), and the NCI NIH R01CA242069 (to L. Cerchietti).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
M.N. Calvo-Vidal: Conceptualization, formal analysis, supervision, investigation, methodology, writing–original draft. N. Zamponi: Formal analysis, investigation, visualization, methodology. J. Krumsiek: Software, formal analysis, visualization. M.A. Stockslager: Investigation. M.V. Revuelta: Software, formal analysis, visualization. J.M. Phillip: Investigation. R. Marullo: Investigation. E. Tikhonova: Software. N. Kotlov: Software. J. Patel: Investigation. S.N. Yang: Investigation. L. Yang: Investigation. T. Taldone: Resources. C. Thieblemont: Resources, patient specimens and data. J.P. Leonard: Resources, patient specimens and data. P. Martin: Resources, patient specimens and data. G. Inghirami: Resources, supervision, patient specimens and data. G. Chiosis: Resources, supervision, writing–review and editing, patient specimens and data. S.R. Manalis: Resources, supervision, investigation. L. Cerchietti: Conceptualization, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing.
References
- 1. Calvo-Vidal MN, Cerchietti L. The metabolism of lymphomas. Curr Opin Hematol 2013;20:345–54. [DOI] [PubMed] [Google Scholar]
- 2. Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 2012;22:547–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vajpayee N, Thakral C, Gopaluni S, Newman N, Gajra A. Activation of mammalian target of rapamycin in diffuse large B-cell lymphoma: a clinicopathological study. Leuk Res 2012;36:1403–9. [DOI] [PubMed] [Google Scholar]
- 4. Dang CV, Lewis BC. Role of oncogenic transcription factor c-Myc in cell cycle regulation, apoptosis and metabolism. J Biomed Sci 1997;4:269–78. [DOI] [PubMed] [Google Scholar]
- 5. Sander S, Calado DP, Srinivasan L, Kochert K, Zhang B, Rosolowski M, et al. Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 2012;22:167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015;17:351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O'Connell JD, Zhao A, Ellington AD, Marcotte EM. Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu Rev Cell Dev Biol 2012;28:89–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmitt DL, An S. Spatial organization of metabolic enzyme complexes in cells. Biochemistry 2017;56:3184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meyer P, Cecchi G, Stolovitzky G. Spatial localization of the first and last enzymes effectively connects active metabolic pathways in bacteria. BMC Syst Biol 2014;8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dhar A, Samiotakis A, Ebbinghaus S, Nienhaus L, Homouz D, Gruebele M, et al. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc Natl Acad Sci U S A 2010;107:17586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu H, Juvekar A, Lyssiotis CA, Lien EC, Albeck JG, Oh D, et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 2016;164:433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shearwin K, Nanhua C, Masters C. Interactions between glycolytic enzymes and cytoskeletal structure–the influence of ionic strength and molecular crowding. Biochem Int 1990;21:53–60. [PubMed] [Google Scholar]
- 13. Volker KW, Reinitz CA, Knull HR. Glycolytic enzymes and assembly of microtubule networks. Comp Biochem Physiol B Biochem Mol Biol 1995;112:503–14. [DOI] [PubMed] [Google Scholar]
- 14. An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc Natl Acad Sci U S A 2010;107:12872–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castellana M, Wilson MZ, Xu Y, Joshi P, Cristea IM, Rabinowitz JD, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol 2014;32:1011–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin M, Fuller GG, Han T, Yao Y, Alessi AF, Freeberg MA, et al. Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep 2017;20:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeon M, Kang HW, An S. A mathematical model for enzyme clustering in glucose metabolism. Sci Rep 2018;8:2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008;320:103–6. [DOI] [PubMed] [Google Scholar]
- 19. Zhao H, Chiaro CR, Zhang L, Smith PB, Chan CY, Pedley AM, et al. Quantitative analysis of purine nucleotides indicates that purinosomes increase de novo purine biosynthesis. J Biol Chem 2015;290:6705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. French JB, Jones SA, Deng H, Pedley AM, Kim D, Chan CY, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science 2016;351:733–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carcamo WC, Calise SJ, von Muhlen CA, Satoh M, Chan EK. Molecular cell biology and immunobiology of mammalian rod/ring structures. Int Rev Cell Mol Biol 2014;308:35–74. [DOI] [PubMed] [Google Scholar]
- 22. Pedley AM, Karras GI, Zhang X, Lindquist S, Benkovic SJ. Role of HSP90 in the regulation of de novo purine biosynthesis. Biochemistry 2018;57:3217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. French JB, Zhao H, An S, Niessen S, Deng Y, Cravatt BF, et al. Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proc Natl Acad Sci U S A 2013;110:2528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joshi S, Wang T, Araujo TLS, Sharma S, Brodsky JL, Chiosis G. Adapting to stress - chaperome networks in cancer. Nat Rev Cancer 2018;18:562–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rodina A, Wang T, Yan P, Gomes ED, Dunphy MP, Pillarsetty N, et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature 2016;538:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pillarsetty N, Jhaveri K, Taldone T, Caldas-Lopes E, Punzalan B, Joshi S, et al. Paradigms for precision medicine in epichaperome cancer therapy. Cancer Cell 2019;36:559–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taldone T, Zatorska D, Patel PD, Zong H, Rodina A, Ahn JH, et al. Design, synthesis, and evaluation of small molecule Hsp90 probes. Bioorg Med Chem 2011;19:2603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goldstein RL, Yang SN, Taldone T, Chang B, Gerecitano J, Elenitoba-Johnson K, et al. Pharmacoproteomics identifies combinatorial therapy targets for diffuse large B cell lymphoma. J Clin Invest 2015;125:4559–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cermak N, Olcum S, Delgado FF, Wasserman SC, Payer KR, AM M, et al. High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat Biotechnol 2016;34:1052–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, et al. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 2007;446:1066–9. [DOI] [PubMed] [Google Scholar]
- 31. Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2.0: interactive pathway explorer. Nucleic Acids Res 2011;39:W412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33:W741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moulick K, Ahn JH, Zong H, Rodina A, Cerchietti L, Gomes DaGama EM, et al. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol 2011;7:818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cerchietti LC, Lopes EC, Yang SN, Hatzi K, Bunting KL, Tsikitas LA, et al. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat Med 2009;15:1369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Culjkovic-Kraljacic B, Fernando TM, Marullo R, Calvo-Vidal N, Verma A, Yang S, et al. Combinatorial targeting of nuclear export and translation of RNA inhibits aggressive B-cell lymphomas. Blood 2016;127:858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leuenberger P, Ganscha S, Kahraman A, Cappelletti V, Boersema PJ, von Mering C, et al. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 2017;355:eaai7825. [DOI] [PubMed] [Google Scholar]
- 37. Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol 2013;31:419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taldone T, Patel PD, Patel M, Patel HJ, Evans CE, Rodina A, et al. Experimental and structural testing module to analyze paralogue-specificity and affinity in the Hsp90 inhibitors series. J Med Chem 2013;56:6803–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ngwa VM, Edwards DN, Philip M, Chen J. Microenvironmental metabolism regulates antitumor immunity. Cancer Res 2019;79:4003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welihinda AA, Kaur M, Raveendran KS, Amento EP. Enhancement of inosine-mediated A2AR signaling through positive allosteric modulation. Cell Signal 2018;42:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dunphy MPS, Pressl C, Pillarsetty N, Grkovski M, Modi S, Jhaveri K, et al. First-in-human trial of epichaperome-targeted PET in patients with cancer. Clin Cancer Res 2020;26:5178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jhaveri KL, Dos Anjos CH, Taldone T, Wang R, Comen E, Fornier M, et al. Measuring tumor epichaperome expression using [(124)I] PU-H71 positron emission tomography as a biomarker of response for PU-H71 plus nab-paclitaxel in HER2-negative metastatic breast cancer. JCO Precis Oncol 2020;4:PO.20.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov 2015;5:1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen L, Papenhausen P, Shao H. The role of c-MYC in B-cell lymphomas: diagnostic and molecular aspects. Genes 2017;8:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ennishi D, Jiang A, Boyle M, Collinge B, Grande BM, Ben-Neriah S, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol 2019;37:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet 2005;37:382–90. [DOI] [PubMed] [Google Scholar]
- 48. Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci 2001;26:597–604. [DOI] [PubMed] [Google Scholar]
- 49. Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 2021;22:196–213. [DOI] [PubMed] [Google Scholar]
- 50. Bereiter-Hahn J, Stubig C, Heymann V. Cell cycle-related changes in F-actin distribution are correlated with glycolytic activity. Exp Cell Res 1995;218:551–60. [DOI] [PubMed] [Google Scholar]
- 51. Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 2016;164:487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 2015;162:1286–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Altman BJ, Dang CV. Normal and cancer cell metabolism: lymphocytes and lymphoma. FEBS J 2012;279:2598–609. [DOI] [PubMed] [Google Scholar]
- 54. Condelli V, Crispo F, Pietrafesa M, Lettini G, Matassa DS, Esposito F, et al. HSP90 molecular chaperones, metabolic rewiring, and epigenetics: impact on tumor progression and perspective for anticancer therapy. Cells 2019;8:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hasko G, Sitkovsky MV, Szabo C. Immunomodulatory and neuroprotective effects of inosine. Trends Pharmacol Sci 2004;25:152–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table 1, Table 2 and Table 3
Data Availability Statement
RNA-sequencing data for WCM patients with DLBCL are available at Gene Expression Omnibus (GEO) with the accession number: GSE145043. The metabolomics dataset of patients with DLBCL and healthy volunteers is available in Supplementary Materials and Methods.