Abstract
Epithelial ovarian carcinomas (OC) are particularly deadly due to intratumoral heterogeneity, resistance to standard-of-care therapies, and poor response to alternative treatments such as immunotherapy. Targeting the OC epigenome with DNA methyltransferase inhibitors (DNMTi) or histone deacetylase inhibitors (HDACi) increases immune signaling and recruits CD8+ T cells and NK cells to fight OC in murine models. This increased immune activity is caused by increased transcription of repetitive elements (RE) that form double-stranded RNA (dsRNA) and trigger an interferon response. To understand which REs are affected by epigenetic therapies in OC, we assessed the effect of DNMTi and HDACi on OC cell lines and patient samples. Subfamily-level (TEtranscripts) and individual locus-level (Telescope) analysis of REs showed that DNMTi treatment upregulated more REs than HDACi treatment. Upregulated REs were predominantly LTR and SINE subfamilies, and SINEs exhibited the greatest loss of DNA methylation upon DNMTi treatment. Cell lines with TP53 mutations exhibited significantly fewer upregulated REs with epigenetic therapy than wild type TP53 cell lines. This observation was validated using isogenic cell lines; the TP53 mutant cell line had significantly higher baseline expression of REs but upregulated fewer upon epigenetic treatment. In addition, p53 activation increased expression of REs in wild type but not mutant cell lines. These data give a comprehensive, genome-wide picture of RE chromatin and transcription-related changes in OC after epigenetic treatment and implicate p53 in RE transcriptional regulation.
Keywords: Ovarian Cancer, Retrotransposon Transcription, DNA methylation, Histone Acetylation, Interferon Signaling, p53
Introduction
About 43% of the human genome is composed of repetitive elements (REs) (1). As REs contain transcription factor binding sites and other regulatory sequences, transcription of these elements is tightly regulated. REs may be expressed in embryonic stem cells but are mostly silenced by DNA methylation and repressive histone modifications in terminally differentiated cells. As part of the global epigenetic dysregulation that normal cells undergo during transformation, REs can lose repressive epigenetic marks, promoting aberrant transcription. Recent work has shown how aberrant RE transcription in cancer, especially from retrotransposons and transposable elements (TEs), can lead to an immune response that promotes anti-tumor immunity (2-6).
There are three main classes of retrotransposons: long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and long terminal repeats (LTRs, also known as endogenous retroviruses or ERVs). About 90% of LTRs have completely lost internal ORFs, leaving only the LTR sequences that cannot transpose (1). Only about 100 of 1.5 million LINEs remain intact enough to retrotranspose (1). Nevertheless, LTR and LINE promoters can alter gene expression (7) and destabilize the genome. As a result, retrotransposon sequences are silenced by epigenetic modifications including DNA methylation and repressive histone modifications (8).
Epigenetic regulation of transcription is disrupted in almost all cancers. This results in genome-wide loss of methylation and local hypermethylation at promoter regions that are normally unmethylated (9). 5-azacytidine (AZA) is a cytosine analogue that inhibits DNA methyltransferases (DNMTi). AZA can reactivate tumor suppressor genes silenced by DNA methylation at their promoters (10). AZA and another DNMTi, 5-aza-2’-deoxycytidine, are approved by the FDA for treatment of myelodysplastic syndrome (10) and AZA is approved for acute myeloid leukemia (11). We (4,12) and others (13-16) have shown that low doses of DNMTis upregulate immune signaling, including the interferon response, cancer/testis antigens (CTAs), and antigen processing and presentation in breast, colon, lung, and OC cell lines (12-16). The activation of the interferon response by DNMTi is caused by upregulation of dsRNA, specifically LTR transcripts that activate the dsRNA sensors TLR3 and MDA5 (4,15). Recent work has demonstrated the role of inverted repeat Alu (IR-Alu, a subgroup of SINEs) elements upregulated by DNMTi treatment (17) that bind to MDA5, triggering interferon signaling (2,17). Interferon signaling due to retrotransposon transcription can also be triggered by inhibitors of histone deacetylases (HDACi) (18) or H3K9 methyltransferases (19). Combining DNMTi and HDACi increased REs in a mouse model of OC (ID8) (20), activating interferon signaling, and recruiting CD8+ T cells to kill the cancer cells (21). This work outlines a clear mechanism by which epigenetic therapies alter the immune microenvironment of cancer through transcriptional regulation of REs. However, it remains unclear which REs are activated by demethylation versus chromatin changes and whether tumor mutational background affects their transcription.
TP53 is a key tumor suppressor and the most commonly mutated gene in human cancers. About 30% of known p53 binding sites in the human genome are in LTRs, and p53 activates transcription of specific LTRs in the human colorectal cancer cell line HCT116 (22). In contrast, p53 can transcriptionally repress REs in fruit flies and zebrafish (23,24). In human cell lines, LINE-1 is silenced by wild type p53 and transcribed in TP53 null cells. Transcription in TP53 null cells is correlated with the loss of the H3K9me3 and H3K27me3 repressive histone marks (25). 90% of TP53 mutations in human cancers cluster in the DNA binding domain and are termed “hotspot mutations”. These hotspot mutations have diverse effects and may prevent p53 binding to canonical targets, instead promoting oncogenic transcription by binding to other loci through interaction with different binding partners (26). It remains unclear how mutant p53 affects retrotransposon expression in cancer, especially in OC where the majority of cases contain a TP53 mutation (27).
To gain an understanding of which REs are affected by specific epigenetic therapies in OC, we assessed the effect of AZA (DNMTi) and ITF-2357 (ITF, an HDAC inhibitor) on OC cell lines and OC patient samples from a DNMTi clinical trial. Subfamily-level (TEtranscripts) and individual locus-level (Telescope) analysis of REs showed that DNMTi treatment upregulated significantly more REs than HDACi treatment, while the combination of DNMTi/HDACi increased RE transcription. Upregulated REs were dominated by the LTR and SINE families and SINEs showed the biggest differences in DNA methylation upon DNMTi treatment. Interestingly, cell lines with TP53 mutations exhibited significantly lower upregulation of REs with epigenetic therapy than wild type TP53 cell lines. We validated this using isogenic cell lines (wild type and mutant TP53) and found that the TP53 mutant cell line had significantly higher baseline expression of REs but upregulated fewer upon epigenetic treatment. Activation of p53 increases expression of REs in wild type but not mutant cell lines, and wild type p53 binds to genomic loci of specific RE families. These data give a comprehensive, genome-wide picture of RE chromatin and transcription changes in OC after epigenetic treatment and implicate p53, a protein mutated in the majority of ovarian cancers, in RE transcriptional regulation.
Materials and Methods
Cell Lines
Human OC cell lines (A2780, Hey, Kuramochi, SKOV3, and TykNu) were kindly given to us by Dr. Stephen Baylin (Johns Hopkins University) and have been verified by STR analysis. The A2780, Hey, SKOV3, and TykNu cell lines were cultured in RPMI 1640 (Corning, 10-104-CV) with 10% Fetal Bovine Serum (X&Y Cell Culture, FBS-500-HI), and 1% penicillin and streptomycin solution (Gibco, 15070063). The Kuramochi cell line was cultured in RPMI 1640 media (Corning, 10-104-CV) with 10% fetal bovine serum (X&Y Cell Culture, FBS-500-HI), 1% penicillin and streptomycin solution (Gibco, 15070063) and 1% non-essential amino acids (Gibco, 11140050). Cell lines were periodically tested for mycoplasma via the Lonza MycoAlert kit.
Drugs and Treatments
Cells were cultured in T75 dishes (Greiner, 658170) and treated with 500 nM 5-azacytidine (AZA, Sigma-Aldrich) or PBS. Media and AZA were replaced each day for five days (Fig. 1B). The cells were then split and allowed to reattach. ITF-2357 was added at a concentration of 100 nM and treatment was continued for two days. The sequential treatment of these cells by these drugs was optimized by Topper et al. (28). The A2780 p53 ChIP-seq samples were treated with 500 nM AZA (Sigma-Aldrich) or PBS for three days and 10 uM Nutlin-3A (Cayman Chemical 18585) or DMSO for 6 hours.
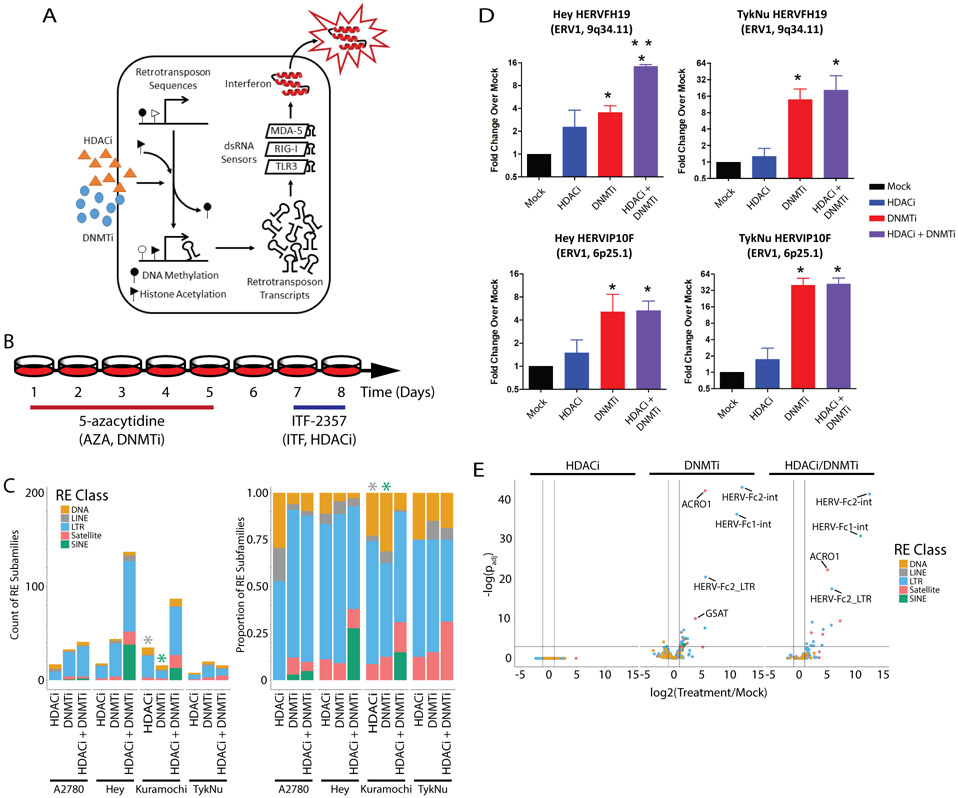
Figure 1: Epigenetic therapies upregulate transcription of repetitive elements.
A) Primary mechanism by which epigenetic therapies upregulate transcription of repetitive elements, especially retrotransposons, to activate the interferon response. B) Treatment scheme for the OC cell lines used. C) Counts of the repetitive element subfamilies upregulated by epigenetic therapies. Each bar represents data from one sample (n=1) for each cell line and treatment combination. Left shows raw counts. Right shows the proportion of counts. Color indicates class of repetitive element. Asterisks indicate enrichment by GSEA (FDR q < 0.05). D) RT-qPCR validation of the upregulation of two LTR loci by epigenetic treatments in the Hey and Kuramochi cell lines. Each RT-qPCR was performed twice in triplicate. Error bars represent the SEM. E) TEtranscripts data volcano plot showing log2(Fold-change) and −log10(DESeq2 padj) for an independent set of three TykNu RNA-seq library replicates. HDACi = ITF. DNMTi = AZA.
RT-qPCR
1 ug of RNA was treated with DNaseI (Thermo #EN0525) for 30 min at 37 C. 1 uL of 50 mM EDTA (4.6 mM final concentration) was added to quench the reaction and the DNase was denatured for 10 min at 65 C. This RNA was used in the RT-qPCR and the remainder was stored at −80 C. The RNA was reverse transcribed using the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit (Cat. No. 4368814) random primers and incubated at 25 C for 10 min, 27 C for 120 min, 85 C for 5 min, followed by a 4 C hold. RT-qPCR was performed with Applied Biosystems SYBR Green on the QuantStudio3 Quantitative reverse transcriptase PCR system. All RT-qPCR primer sequences are listed in Table S1.
CRISPR/Cas9 Genome Engineering
Hey (TP53 wild type) cells were electroporated with a non-targeting control gRNA or a gRNA targeting TP53 at amino acid 175 previously screened to have the highest editing efficiency. RNPs were formed by complexing crRNA and tracrRNA and subsequently adding recombinant Cas9 V3 (IDT) according to the manufacturer's recommendations. For generating the R175H point mutation, HDR templates were designed and synthesized as ssODNs ranging from 75-150bp in length containing the desired base pair change together with a silent point mutation at amino acid 175 as well as a mutated PAM site and an additional silent point mutation to allow for analysis using RFLP (restriction fragment length polymorphism). Multiple polyclonal lines were generated from which single-cell clones were subsequently expanded and tested for TP53 mutation via CRISPResso (29) and functional analyses (Nutlin-3A treatment followed by Western blot and qRT-PCR of target genes (Fig. 7A,B)).
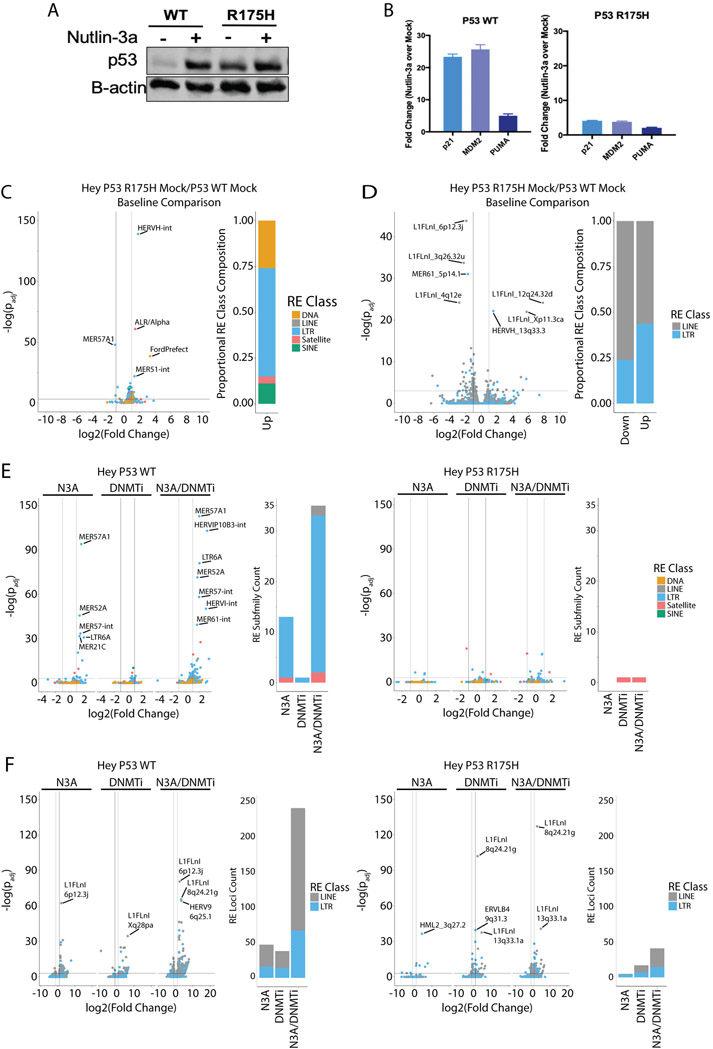
Figure 7: Mutant TP53 changes baseline expression and DNMTi induction of repetitive elements in OC cell lines.
A) P53 western blots from CRISPR/Cas9-edited TP53 wild type (HC2) and R175H mutant (HH23) cell lines expanded from single clones. B) RNA was isolated from the cells in A) and qRT-PCR performed for P53 target genes. C) TEtranscripts volcano plot showing the comparison between the TP53 Mutant and TP53 WT Hey-derived lines. Bar plot at right indicates the class composition of the REs that are significantly upregulated in the TP53 R175H cell line with a greater than two-fold change in expression. D) Telescope volcano plot showing the comparison between the TP53 Mutant and TP53 WT Hey-derived lines. Bar plot at right indicates the class composition of the REs that are significantly differentially expressed with a greater than two-fold change in expression. E) TEtranscripts volcano plots showing REs upregulated by epigenetic therapy in the TP53 WT line (HC2, left) and the R175H mutant line (HH23, right). Bar plots show the count of elements that are significantly upregulated with greater than two-fold change in expression to emphasize the differences between the TP53 WT and mutant lines. F) Same as in E) except the data are from Telescope analysis of TP53 WT and mutant cell lines. DNMTi = AZA.
Western Blot
Cells were lysed in RIPA buffer (Pierce, 89900) with 1X protease and phosphatase inhibitor (Pierce, A32961). Lysates were sonicated at 4°C in a water bath Bioruptor™ (Diagenode) for 8 minutes (8 cycles of 30s on, 30s off). Next, samples were centrifuged at 4°C, 10,000xg for 10 minutes to remove cellular debris. Protein concentration was determined according to the Pierce BCA Protein Assay Kit protocol (Thermo Fisher Scientific, 23225). Samples were mixed with NuPAGE LDS 4x loading gel (NP0007) and NuPAGE 10x reducing agent (NP0009) then placed on a heating block at 100°C. Samples were loaded into a 4-20% (BioRad, 4561093) and transferred to LF PVDF (BioRad, 170-4274). Membranes were blocked with LI-COR Biosciences (Lincoln, Nebraska, USA) Odyssey Blocking Buffer (927-40100) for 2 hours at room temperature then incubated overnight at 4°C with the primary antibody, 1:1000 p53 (rabbit, Bethyl A300-247). After the overnight incubation, Beta-actin (mouse, Sigma A5441) was added, 1:3000, at room temperature for 20 minutes. Proteins were detected using the Azure Biosystems Imaging System c600. Processing of images was performed using the LI-COR Biosciences Image Studio software. The secondary antibodies used were AzureSpectra700 AC2128 and AzureSpectra800 AC2135.
p53 ChIP-seq Library Preparation
ChIP-seq library preparation was adapted from the Active Motif ChIP-IT High Sensitivity Kit protocol (Active Motif, 53040). A2780 cells were grown and treated in 150cm dishes until 80% confluency, aiming for 15 million cells per treatment prior to crosslinking. Cells were cross-linked for 10 minutes at room temperature. The samples were sheared using 2 μL MNase (NEB, M0247S) for 10 minutes, then briefly sonicated 4 cycles (30secs on/30secs off) at 4°C. Samples were then centrifuged and the supernatant was used as the input and for the downstream immunoprecipitation. The immunoprecipitation was performed following the manufacturer’s protocol of the Active Motif Kit, using 4 μL of the p53 antibody (Bethyl Laboratories, A300-247A-M). DNA was purified and used for qPCR analysis to verify enrichment prior to ChIP-Seq. ChIP-seq was completed using the NEBNext Ultra II DNA Library Prep (NEB, E7103S) with multiplex oligos for barcoding (NEB, E7335S).
RNA-seq Library Preparation
Following treatment, total RNA was extracted using Trizol (Thermo Scientific 15596026). Ribosomal RNA was depleted using Ribozero (Illumina). RNA sequencing libraries were prepared using the ScriptSeq v2 RNA-seq Library Preparation kit (SSV21106). Indexed libraries were sent to the Washington University in St. Louis Center for Genome Sciences for 75 bp, paired-end sequencing. The triplicate TykNu libraries were prepared for sequencing by depleting rRNA from 1 ug of total RNA with the NEBNext rRNA Depletion Kit for Human (New England BioLabs, E6310). Libraries were then prepared by following the manufacturer’s protocol for the Illumina TruSeq® Stranded Total RNA Library Prep Human/Mouse/Rat kit (P/N 20020596). Custom 10 bp UDI TruSeq-Compatible Duplex Y Adapters from IDT were to index the libraries (IDT10_UDI_1 through IDT10_UDI_15). Final libraries were sent to the Washington University in St. Louis Center for Genome Sciences and combined into an equimolar pool for 2x150bp sequencing on an Illumina NovaSeq S4 (target read depth of 100 million). TP53 wild type (isogenic Hey-derived, CRISPR-edited control line designated HC2), and TP53 mutant (isogenic Hey-derived, CRISPR-edited R175H mutant line designated HH23) libraries were produced from 750 ng of total RNA with a minimum RIN score of 7.0. This RNA was prepared for sequencing following the manufacturer's protocol for the Illumina TruSeq® Stranded Total RNA Library Prep Human/Mouse/Rat kit (P/N 20020596) and TruSeq RNA Single Indexes Set A and B (P/N 20020492 and 20020493). Final libraries were separated into two sets of 12 and each set was combined into an equimolar pool for sequencing on an Illumina NextSeq 500 using two High Output 150 cycle v2.5 kits (P/N 20024907) and PhiX Control v3 (P/N FC-110-3001) spike-in of 1%.
ATAC-seq Library Preparation
ATAC-Seq was performed according to Corces et al. (30). Following treatment, DNA was extracted using the Nextera DNA Library Prep Kit (FC-121-1030). The libraries were prepared according to Corces et al. (30). Indexed libraries were sent to the Washington University in St. Louis Center for Genome Sciences for 75 bp, paired-end sequencing on an Illumina NextSeq500.
MeDIP-seq and MRE-seq Library Preparation
Following treatment and DNA extraction with phenol/chloroform, DNA was sent to the Ting Wang lab at Washington University in St. Louis. Library preparation occurred as described by Xing et al. (31). For MRE-seq, only the HpaII, SsiI, Hin6I, and HpyCH4IV restriction enzymes were used. The finished libraries were sequenced at the Center for Genome Sciences at Washington University on an Illumina NextSeq500.
Annotation Files
The hg38 reference sequence corresponded to the initial release without patches, GCA_000001405.15. The sequences were downloaded from ftp://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/analysisSet/* on April 5, 2018.
The GENCODEv21 genomic feature annotations downloaded from ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_21/gencode.v21.chr_patch_hapl_scaff.annotation.gtf.gz on June 4, 2018.
The RepeatMasker annotation (32) used here is: hg38 - Dec 2013 - RepeatMasker open-4.0.5 - Repeat Library 20140131 and can be accessed here http://www.repeatmasker.org/species/hg.html. The RepeatMasker table was reformatted into GTF and BED files for use in subsequent analysis.
TEtranscripts RNA-seq Analysis
The sequence quality of the FASTQ files was assessed with FastQC. Reads were trimmed and adapters were removed using cutadapt using the --minimum-length 1 and -q 20 flags. The adapter sequences were -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC and -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT. Trimmed reads were aligned to hg38 using the following flags: --sjdbOverhang 100 --winAnchorMultimapNmax 200 --outFilterMultimapNmax 100. For libraries prepared by the ScriptSeq v2 RNA-seq, TEtranscripts was run on the STAR output with the following flags: --mode multi --stranded yes. For later triplicate libraries prepared by the Illumina TruSeq® Stranded Total RNA Library Prep Human/Mouse/Rat kit, TEtranscripts was run on the STAR output with the following flags: --mode multi --stranded reverse. The GENCODEv21 and RepeatMasker GTF annotation files were supplied. The log2(Fold Change) values output by DESeq were used in subsequent analyses. See Jin et al. for further reference (33). Library quality metrics are in Table S2.
Telescope RNA-seq Analysis
Telescope analysis was performed on the same aligned reads files as TEtranscripts. Telescope was installed and used according to the guidelines here: https://github.com/mlbendall/telescope. Before starting, miniconda was installed with the bioconda and conda-forge channels. Then a conda environment specifically for this task was created: conda create -n telescope_env python=3.6 future pyyaml cython=0.29.7 numpy=1.16.3 scipy=1.2.1 pysam=0.15.2 htslib=1.9 intervaltree=3.0.2. In this environment, the “telescope assign” command was used to quantify RE expression. The output *report.tsv files were combined for processing with DESeq2, and the log2(Fold Change) values were used in subsequent analysis. For further information, see Bendall et al. (34). Library quality metrics are in Table S2.
TCGA RNA-seq Analysis
TCGA RNA-seq counts from HTSeq were downloaded from the GDC Data Portal (https://portal.gdc.cancer.gov/). Differential expression was calculated by DESeq2.
ATAC-seq Analysis
The sequence quality of the FASTQ files was assessed with FastQC (0.11.5). Reads were trimmed and adapters were removed using cutadapt (1.16) using the --minimum-length 1 and -q 20 flags. The adapter sequences were -a CTGTCTCTTATACACATCT and CTGTCTCTTATACACATCT. Trimmed reads were aligned to hg38 with Bowtie2 (2.2.9) using the -X 2000 flag. To create input for HMMRATAC, the Bowtie2 output was converted to a BAM file, sorted, and indexed with samtools (1.3.1) view, sort, and index commands on the default settings. MACS2 callpeak (2.1.1.20160309) was used to create a peaks file on the standard settings. The bedGraph file output by MACS2 was sorted using the bash “sort -k1,1 -k2,2n” command then converted to a bigWig with bedGraphToBigWig. The HMMRATAC_v1.2.1 Java executable was run with the following flags: -m 75,200,400,600 --window 1250000 --bedgraph true -u 20 -l 10 -z 100. The peaks were filtered for a minimum score of 10 (i.e. at least 10 reads map to the open region). Then bedtools intersect (2.26.0) was used to remove peaks that had an 85% reciprocal overlap with peaks from the mock treated sample. These treatment specific peaks were used in subsequent analyses. For further reference, see Tarbell and Liu (35). Library quality metrics are in Table S2.
methylMnM and methylCRF Analysis
The sequence quality of the FASTQ files was assessed with FastQC (0.11.5). Reads were trimmed and adapters were removed using cutadapt (1.16) using the -q 20 flags. The adapter sequences were -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC and -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT. The reads were aligned to hg38 using bwa mem (0.7.12) on default settings. MeDIP and MRE data quality was further assessed using methylQA (0.2.1). Reference files for hg19 are publicly available. We made the necessary hg38 reference files for the methylMnM analysis using custom scripts provided by the Wang lab. Rscripts (R version 3.4.2) were used to further process the files and output DMRs (methylMnM) as well as predict methylation levels (methylCRF). For details, follow the protocol described in Xing et al. 2018 (31). Library quality metrics are in Table S2.
ChIP-seq Analysis
ChIP-seq reads were examined for gross problems with FastQC. Adapter removal and trimming were performed with cutadapt using this adapter sequence: -a AGATCGGAAGAGCACACGTCTGAACTCCAGT as well as the -q 20 and –minimum-length 1 flags. Reads were aligned to the genome with Bowtie2, and the alignments were quality checked with ChIPQC. Duplicate reads were marked with GATK MarkDuplicates (v4.0.8.0), and peaks were called with MACS2. Peaks consistent across replicates were identified with the ENCODE IDR pipeline (https://github.com/kundajelab/idr) using an IDR cutoff of 0.05. Library quality metrics are in Table S2.
Mehdipour et al. IR-Alu Analysis
The list of IR-Alus reported by Mehdipour et al. was shared with us by the Daniel de Carvalho laboratory at the University of Toronto and the liftOver tool was used to convert the hg19 coordinates to hg38. The methylCRF data in bigWig format was used via the deepTools computeMatrix and plotProfile tools to create metaplots of the methylation values over these IR-Alus.
Statistics
Unless otherwise specified, all statistics were performed in R. The RT-qPCR statistics were run in Prism. When not performed as part of TEtranscripts or Telescope, RNA-seq expression values were called by DESeq2 (36). Venn diagrams were created using Vennerable, except for the nested Venn diagrams in Figure 3 that were created with the eulerr package. Multi-set significance was calculated with the SuperExactTest package (37). For enrichment calculations involving lists of differentially expressed REs, we used gene set enrichment analysis preranked analysis on the RE log2(Fold Change) values (38). For enrichment calculations involving overlap of DMR, ATAC-seq peak, or ChIP-seq peak regions with REs, we used the regioneR package, running 1000 permutations (39).
Figure 3: Epigenetic therapies increase transcription of specific LTR and LINE loci.
A) Distribution of the transcription fold-change for individual LTR and LINE loci as determined by Telescope. B) Telescope data volcano plot showing log2(Fold-change) and −log10(DESeq2 padj) for an independent set of three TykNu RNA-seq library replicates. C) Count of upregulated LTR and LINE loci. The innermost group was present in all four OC lines and each circle outward indicates upregulation in one less line. This data is also presented in bar chart form in Supplemental Figure 3. D) Cluster of repetitive element loci that lose methylation over the 5 kb upstream of the start coordinate and are upregulated. Each plot is centered on the LTR or LINE start coordinate, and each dot represents differential methylation at a single CpG (indicated by blue tick marks). Blue text indicates the locus ID. The number in red text indicates the upregulation fold change. Additional WIMSi clusters can be found in Figure S5. E) Metaplots of average methylation values covering the IR-Alus reported by Mehdipour et al. Graphs present mean methylation and shaded areas indicate the standard deviation. HDACi = ITF. DNMTi = AZA.
Availability of Data and Materials
The OC cell line RNA-seq, methylMnM, methylCRF, ChIP-seq, and ATAC-seq data are available in GEO, accession GSE182430. The trial data from the Fang et al. study (40) is available in GEO, accession GSE102120. TCGA data was accessed via the NIH GDC Data Portal (https://portal.gdc.cancer.gov/). Custom scripts from our laboratory used in the analyses presented here can be found on the Chiappinelli Lab Github page (https://github.com/Chiappinelli-Lab/Ovarian-Cancer-RE-Analysis-Scripts).
Results
Inhibiting DNA Methylation Drives Global Repetitive Element Expression
Epigenetic therapy upregulates expression of a subset of LTRs to activate an interferon response in OC (Fig. 1A) (4). To identify other REs that change expression following epigenetic therapy, we treated the A2780, Hey, Kuramochi, and TykNu OC cell lines with 5-azacytidine (AZA, a DNMTi), ITF-2357 (ITF, an HDACi), or a combination of the two (Fig. 1B). RNA-seq libraries from these samples were analyzed by TEtranscripts (33). This tool analyzes subfamily-level repetitive element (RE) expression by combining counts from all loci of a given RE subfamily. Both DNMTi and DNMTi/HDACi treatments increased interferon-stimulated gene (ISG) expression (Fig. S1), as expected based on previous analysis by qRT-PCR in lung cancer and OC (28,41).
409 repetitive element subfamilies of all major classes (LTR, LINE, SINE, DNA, and satellite repeats) were differentially expressed following treatment by DNMTi and/or HDACi (Fig. 1C, Fig. S2A-C). The expression levels of several individual, upregulated retrotransposons, where primer design was possible, were validated by RT-qPCR (Fig. 1D). LINE and SINE transposon classes were significantly enriched in gene set enrichment analysis as represented by the asterisks in Fig. 1C. Though not statistically enriched, LTR retrotransposons were the most frequently upregulated transposon type and have been highlighted as key initiators of interferon signaling downstream of DNMTi treatment (4,15) as have inverted repeat Alu (IR-Alu, SINE) elements (17). DNMTi upregulated more REs than HDACi and the combination of HDACi with DNMTi treatment upregulated a greater number of SINE elements than either treatment alone (Fig. 1C).
While REs were upregulated in all OC cell lines by DNMTi and DNMTi/HDACi treatment, few RE subfamilies were commonly differentially expressed in any three or more of the four OC cell lines (Table S3). HERV9-int was the only LTR subfamily upregulated in all four cell lines by the DNMTi/HDACi combination treatment (Table S3). Several other LTR, LINE, and satellite repetitive elements were upregulated in any three cell lines (Table S3-5). Though few REs were upregulated across multiple lines, the overlap of these REs was nevertheless statistically significant and unlikely to occur by chance (Fig. S2D). Further analysis of RE families after epigenetic treatment in the TykNu cell line confirmed that HDACi alone had very little effect while DNMTi drove RE transcription increases both alone and in combination with HDACi. Notably, many of the RE subfamilies that underwent DNMTi-induced transcription were LTRs (Fig. 1E).
Epigenetic Therapies Induce Correlated Epigenetic and Transcriptomic Changes
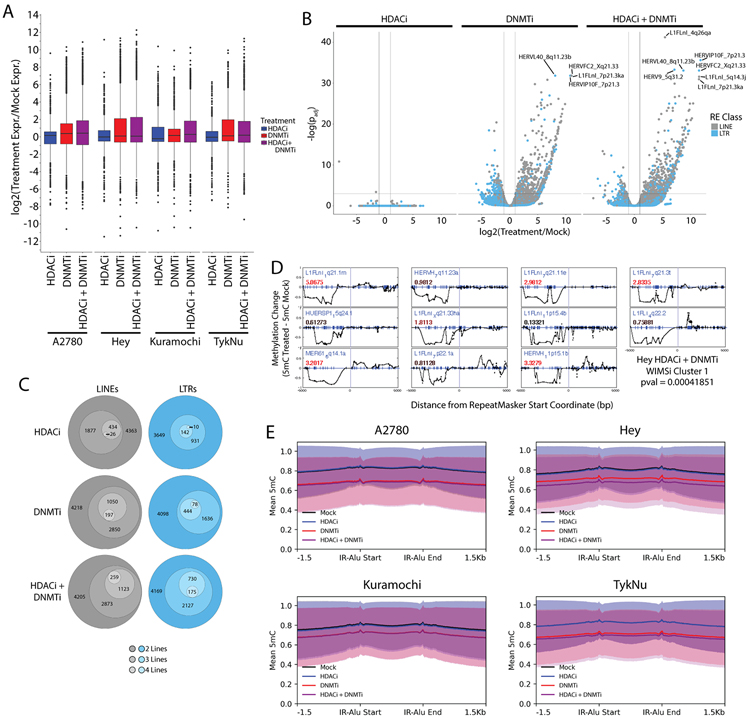
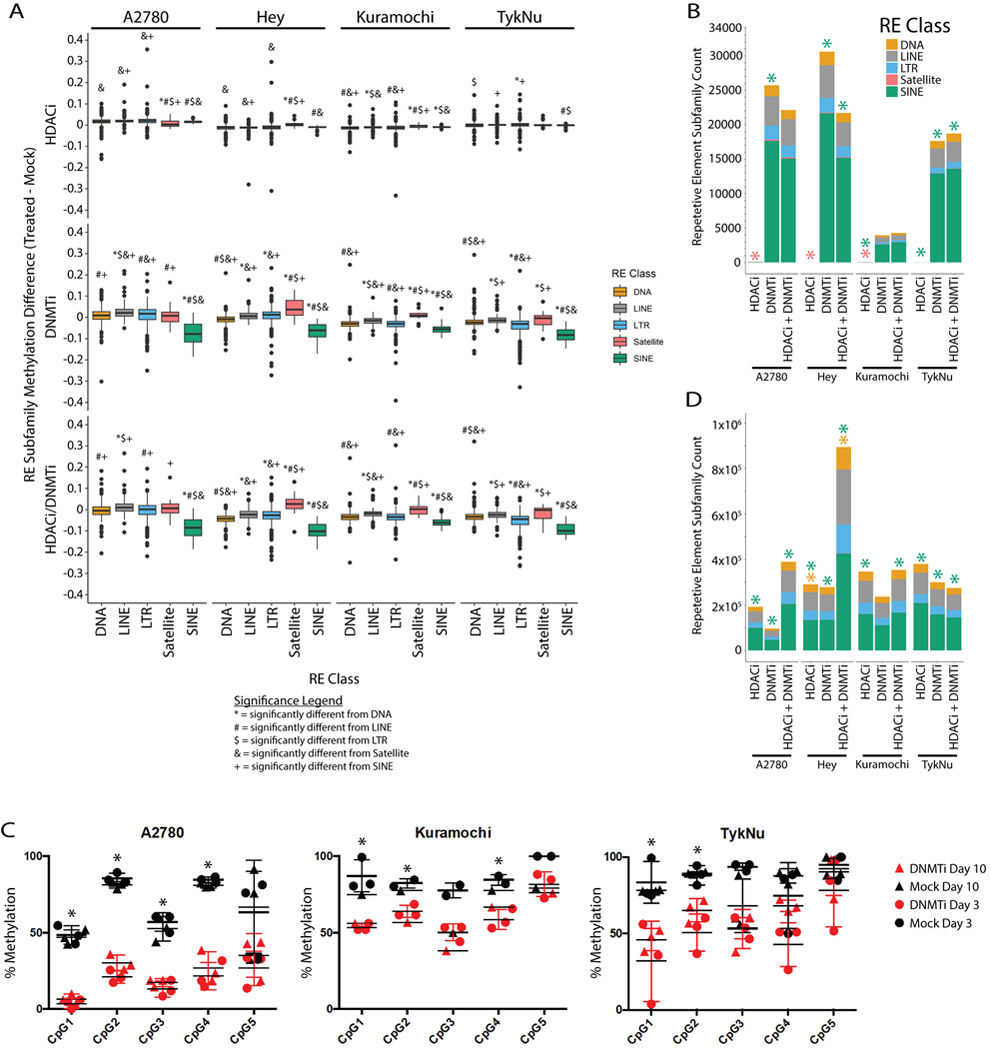
We next explored whether differential methylation or chromatin accessibility were responsible for the differential expression of RE subfamilies. We utilized MeDIP-seq and MRE-seq (31) followed by analysis with the methylCRF algorithm (31) to determine methylation at each CpG. This allowed us to calculate the average methylation change of each RE element subfamily or class (Fig. 2A). As expected, HDACi had little effect on DNA methylation while DNMTi alone or the DNMTi/HDACi combination drove methylation loss in all four cell lines. We observed the most significant demethylation at SINE elements (Fig. 2A, 2B). We validated DNMTi-induced methylation loss at the HERV-Fc2 5’ LTR by pyrosequencing analysis (Fig. 2C). This subfamily was one of the most significantly upregulated by DNMTi and DNMTi/HDACi in Fig. 1E.
Figure 2: Retrotransposons, especially SINE elements, are demethylated by DNMTi treatment.
A) Distribution in the average methylation difference for each repetitive element class. Methylation values are averaged across the entire repetitive element. The data for each cell line and treatment combination represent a single methylCRF library. Symbols indicate significance in a Kruskal-Wallis test and subsequent pairwise Wilcox test with Benjamini and Hochberg correction (p < 0.05). Individual symbols indicate a pairwise difference from a specific class (see legend). B) Counts of each repetitive element subfamily that overlap a differentially methylated region. Asterisks indicate significant enrichment of an RE class as determined by the regioneR package. C) Pyrosequencing percent methylation measurements at five CpGs in the HERV-Fc2 5’ LTR from three different cell lines. Asterisks indicate significant difference between the mock and AZA treated samples by t-test (p < 0.05). Error bars represent the SEM. D) Same as B except the regions used in the enrichment calculations were the ATAC-seq peaks called by HMMR-ATAC. HDACi = ITF. DNMTi = AZA.
To better understand which RE classes were most affected by DNMTi and HDACi treatments, we calculated the enrichment of these repetitive elements within the regions of epigenetic change (Fig. 2B,D). For DNA methylation, we called differentially methylated regions (DMRs) between the control and treated samples using methylMnM (31). We called regions of open chromatin from our ATAC-seq data using HMMRATAC (35). Though SINE elements had greater loss of methylation (Fig. 2A) and were enriched in both DMRs and ATAC-seq peaks (Fig. 2B,D), they were less affected at the transcriptional level (Fig. 1). From this data we infer that SINE elements are the primary targets for the loss of methylation and gain of chromatin accessibility upon DNMTi treatment but that these changes are not strongly correlated with transcription from SINE elements.
Epigenetic Therapy Significantly Upregulates Individual Repetitive Element Loci
Correlating the broad epigenetic and expression changes at the subfamily level is problematic because data summarizing the effect at all loci mutes the signal from individual retrotransposon loci. To address this, we used the tool Telescope (34) to analyze LTR and LINE1 expression changes from individual loci. Telescope considers a curated set of 14,968 (approximately 1.9% of 771,683 total) LTR loci and 13,545 (approximately 0.8% of 1,609,790 total) LINE1 loci that slightly enrich for more intact, younger elements (42). We found that individual LTR and LINE1 loci are upregulated, especially by DNMTi treatment (Fig. 3A,B, Fig. S3A) and several thousand LTR and LINE loci are upregulated by DNMTi and HDACi in OC cell lines (Fig. 3C, Table S6-7). The number of upregulated elements common to all four cell lines is lowest for HDACi (26 LINEs, 142 LTRs) and higher for DNMTi (197 LINEs, 444 LTRs) and DNMTi/HDACi (259 LINEs, 175 LTRs) (Fig. 3C, Fig. S3B-E). Again, the overlap of all upregulated Telescope REs was small but statistically significant across cell lines (Fig. S3F).
To determine which REs lost methylation and gained expression, we used the WIMSi technique (43) with a list of LTR and LINE sites from Telescope. WIMSi calculates the difference in methylation between two samples at each CpG within 5 kb of a gene’s transcription start site and interpolates a curve that represents the methylation pattern. These curves are clustered by similarity and a statistical test is applied to find clusters where more genes are coordinately up- or down-regulated than expected by chance (43). We identified several clusters of REs that enriched for loci with similar methylation and expression changes (Fig. 3D, S4A-E). The strongest cluster shows a correlation between upstream hypomethylation and upregulation of LTR and LINE loci upon treatment with DNMTi and HDACi (Fig. 3D). Each plot is centered on the LTR or LINE start coordinate, and each dot represents the difference in methylation at a single CpG (indicated by blue tick marks). Positive values indicate increased DNA methylation in the treated sample compared to the control and negative values indicate decreased DNA methylation in the treated sample compared to the control. Blue text indicates the locus ID. The number indicates the expression fold change; red text represents RE upregulation, and green text indicates RE downregulation. The correlated changes in Fig. 3D are consistent with the known promoter activity of LTR and LINE sequences in general, though promoter activity at these specific loci is not guaranteed. The other clusters (Fig. S4A-E) are weaker and suggest variability in the epigenetic changes induced by DNMTi and HDACi that hinders the clear identification of a good anchor point to use for this correlation (Fig. S4F,G).
Besides decreases in methylation at specific RE loci, we observe a significant demethylation of inverted repeat Alu elements throughout the genome (Fig. 3E). It was recently shown that DNMTi demethylation of orphan, often intronic CpG islands led to upregulation of inverted repeat Alu elements in colorectal cancer. This study found that the large majority of REs that bind to MDA5 and trigger immune signaling are IR-Alus, though LTRs and other REs were also present (17). These elements, which we find are significantly upregulated by DNMTi treatment (Fig. 1E), are also demethylated by DNMTi treatment in OC cell lines (Fig. 3E).
REs Upregulated in by DNMTi in OC Cell Lines are also Upregulated by DNMTi in OC Patient Samples
Having identified specific REs upregulated by epigenetic therapies in OC cell lines, we expanded our analysis to OC patient samples and found similar results. We analyzed patient samples from a randomized controlled trial testing the combination of guadecitabine (DNMTi) and carboplatin in OC patients (40). Guadecitabine is a dinucleotide that resists cytosine deaminase degradation (44) and releases decitabine to inhibit DNMTs by the same mechanism as AZA. This trial demonstrated that DNMTi can reverse carboplatin resistance in OC patients by undoing methylation-induced gene silencing. Fang et al. included a total of 98 patients and produced RNA-seq libraries from 40 of them (40) with post-treatment RNA-seq libraries for eight patients. Our analysis contains an “all” category where all pretreatment (n = 40 patients, 75 samples) and all posttreatment (n = 8 of the 40 patients, 17 samples) samples are combined as well as analysis of the duplicate pre/post-treatment samples for eight patients individually. These latter samples are designated by a four-digit code (i.e. 0203). 980 RE subfamilies and 20,744 individual RE loci were differentially expressed as analyzed by TEtranscripts (Fig. S5A,B, Table S8-9) and Telescope (Fig. S5C,D, Table S10-11), respectively. The overall distribution of RE classes was similar to our cell line models: LTRs dominate the upregulated RE subfamilies (Fig. S5A-D). The OC patients, like the cell lines, demonstrated variability in REs induced (Fig. S5E,F). Only a single subfamily was upregulated in five or more of the eight paired samples when analyzed by TEtranscripts: 5S rRNA. However, there was significant overlap between the REs upregulated by DNMTi treatment in any OC cell line and the REs upregulated by DNMTi treatment in any OC patient sample, both at the subfamily (TEtranscripts) and individual locus (Telescope) levels (Fig. S5G). These data show that 1) OC cell line responses mimic patient responses to DNMTi, showing upregulation of LTRs, and 2) any given RE may be targeted by epigenetic therapies and contribute to the immune response, but it is not common for the same RE subfamilies and/or loci be targeted in multiple patients and/or cell lines (Table S5, 7, 9, 11).
Influence of TP53 on Retrotransposon Expression
To further understand the variability of RE upregulation in OC lines and patient samples, we examined whether TP53 mutational status affected RE expression after epigenetic therapy. Approximately 95% of high grade serous OC tumors harbor TP53 mutations (27). Several studies have linked p53 with the regulation of retrotransposon transcription (22,45). About 30% of p53 binding sites are located in LTR sequences, and p53 binding upregulates these LTRs (22). When we compare WT to mutant TP53 OC cell lines, the mutant cell lines exhibit significantly less upregulation of REs with any epigenetic therapy compared to the wild type cell lines at both the class/subfamily level TEtranscripts (Fig. 4A-D, S6A) and the locus-level Telescope data (Fig 4E, Fig. S6B,C). Though the differences shown in Figure 4A are small, they are meant to show the role of wild type p53 in promoting RE upregulation when all RE classes/sub-families are considered in aggregate. This relationship is highlighted in subsequent plots for specific sub-families/loci in Figure 4B-E and is especially prominent for SINEs (Fig. 4B).
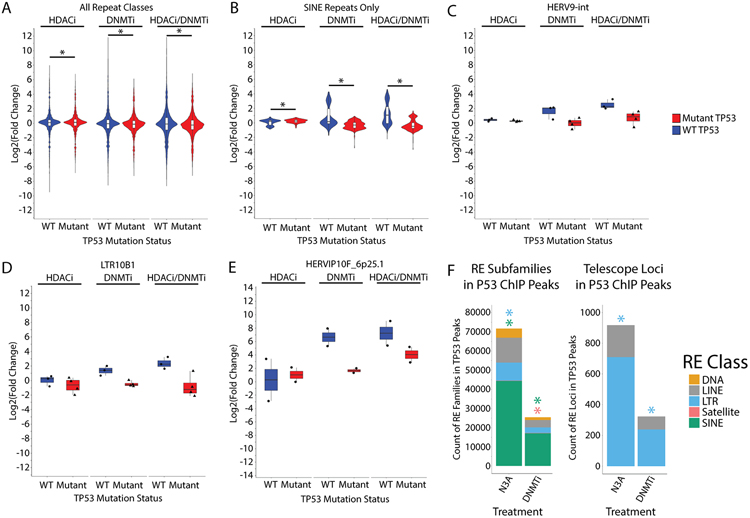
Figure 4: TP53 mutation status and binding sites affect repetitive element expression in OV cell lines.
A) Distribution of transcription fold-change for RE subfamilies analyzed by TEtranscripts. Samples are sub-divided by TP53 mutation status and individual plots are created for repetitive element classes. WT cell lines are A2780 and Hey. Mutant cell lines are Kuramochi and TykNu. Asterisks indicate p < 0.05 by t-test. B) Same as A) except only data from SINE subfamilies is presented. Asterisks indicate p < 0.05 by t-test. C) Same as A) except only data for the HERV9-int subfamily is presented. D) Same as A) except only data for the LTR10B1 subfamily is presented. E) Distribution of Telescope fold-change for the HERVIP10F_6q25.1 locus. F) Count of RE subfamilies (left) or individual LTR or LINE loci (right) that overlap a P53 ChIP-seq peak in A2780. Those significantly enriched (p < 0.05) by regioneR R package are indicated by asterisks. HDACi = ITF. DNMTi = AZA.
To learn more about which REs are regulated by direct wild type p53 binding, we performed p53 ChIP-seq on the TP53 wild type A2780 cell line (Fig. 4F, S7A-F). We treated the cells with Nutlin-3A (N3A) or AZA (DNMTi). Nutlin-3A activates wild type p53 by binding to MDM2, a negative regulator of p53, to inhibit its repressive activity (46). Nutlin-3A induced p53 peaks at RE families, significantly enriched for LTRs and SINEs (Fig. 4F, Table S12). Surprisingly, DNMTi treatment had a similar effect to Nutlin-3A, inducing p53 binding at all major classes of REs including the LTR, LINE, and SINE retrotransposons, though not to the same extent, and only significantly enriching for SINEs (Fig. 4F). At the individual locus level, LTRs were significantly enriched in p53 ChIP-Seq data following Nutlin-3A or DNMTi treatment (Fig. 4F). From these data, we conclude that p53 binding to REs follows DNMTi treatment and affects RE transcription.
Having observed an effect of p53 on RE transcription, we expanded our analysis to analyze RE expression and TP53 mutations in The Cancer Genome Atlas OC RNA-seq data (27). TCGA includes 372 total RNA-seq data sets with matching mutation data. We compared TP53 wild type and mutant samples, all untreated. Interestingly, TP53 mutant cancers tended to express more individual LTR and LINE elements than their WT counterparts (Fig. 5A-C). Mutant p53 status was also associated with an increase in lymphoid infiltration (though non-significant) based on xCell analysis of RNA-seq data (47) (Fig. S8A,B) as well as ISG upregulation (Fig. S8C). Of the 308 RE loci that were upregulated in mutant TCGA OC samples (compared to wild type samples), 115 were also upregulated in the mock treated TP53 mutant lines (Kuramochi and TykNu) compared to the mock treated TP53 wild type lines (A2780 and Hey) (Fig. S8D). Interestingly, only 14 of the 308 were also upregulated by epigenetic therapies. These data suggest that TP53 mutant OCs have baseline higher levels of specific REs, separate from those targeted by epigenetic therapies.
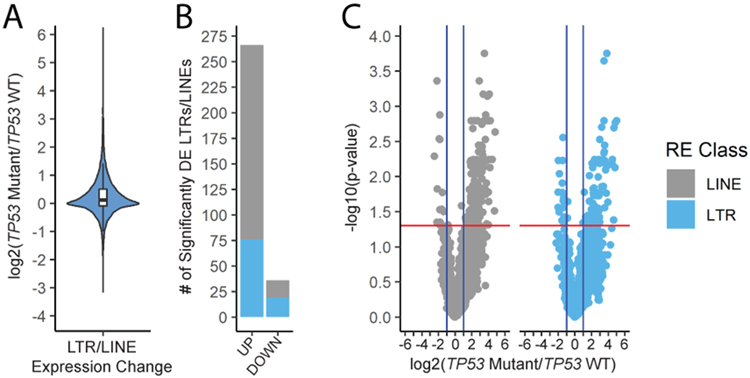
Figure 5: TP53 mutation status affects repetitive element expression in TCGA samples.
A) Distribution of the fold changes for individual LTR and LINE loci. For A-C, the data are TCGA OC samples: WT n=17, Mutant n=224. B) Count of LTR and LINE loci that are upregulated or downregulated in samples with mutant TP53. C) Volcano plots showing significance and magnitude of the expression changes in TCGA patient samples.
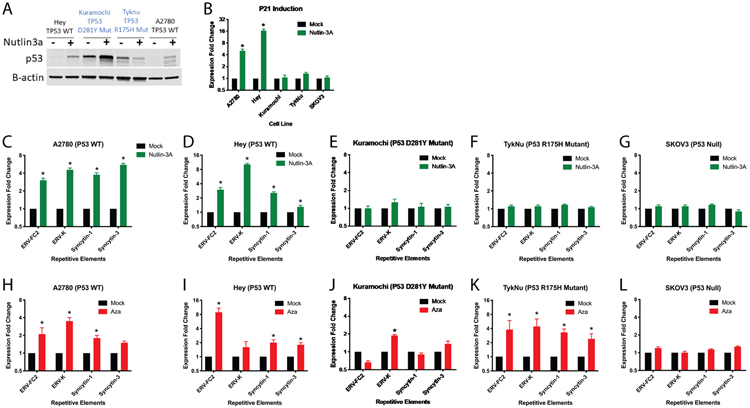
To validate our observed induction of RE expression by p53, we performed targeted experiments in OC cell lines. These included the TP53 wild type A2780 and Hey lines, the TP53 mutant Kuramochi (p.D281Y, c.841G>T) and TykNu (pR175H, c.524G>A) lines, and SKOV3, which contains a nonsense mutation that prevents expression of the p53 protein (48). As expected, Nutlin-3A treatment increased p53 protein levels in the A2780 and Hey wild type cell lines, but not the TykNu and Kuramochi mutant cell lines (Fig. 6A) and increased transcript levels of CDKN1A (P21) (Fig. 6B), an important cell cycle regulator and downstream transcriptional target of p53, in only the WT cell lines. Nutlin-3A treatment increased LTR (ERV-FC2 env, ERV-K env, Syncytin-1 (ERV-W1) env, and Syncytin-3 (ERV-P(b) env) expression in TP53 wild type cell lines (Fig. 6C,D) but not in TP53 null or mutant cell lines (Fig. 6E-G). ERV-FC2, Syncytin-1, and Syncytin-3 were previously shown to be increased by DNMTi in OC cell lines (4) and the FC2 subfamily was one of the most significantly upregulated by DNMTi and DNMTi/HDACi the OC cell lines profiled here (Fig. 1E). DNMTi upregulates LTR loci in a pattern similar to Nutlin-3A (Fig. 6H-L). However, the p53 hotspot mutant cell lines maintain upregulation of REs with DNMTi (Fig. 6J,K).
Figure 6: Effects of TP53 on repetitive element expression.
A) Immunoblot of P53 in OC cell lines treated with Nutlin-3a or control, 24 hours after Nutlin-3a treatment. B-actin was used as a loading control. Cell lines were treated with Nutlin-3a and RNA was extracted 24hours after treatment. B) qRT-PCR was performed for the CDKN1A (P21) transcriptional target of P53 in RNA from the A2780, Hey, and SKOV3 cell lines. C-G) qRT-PCR was performed for RE transcription in C) A2780 TP53 wild type, D) Hey TP53 wild type, E), Kuramochi TP53 mutant, F) TykNu TP53 R175H mutant, and G) SKOV3 TP53 null cell lines. H-L) TP53 WT, R175H, or null cell lines were treated with AZA and LTRs measured by qRT-PCR. H) A2780 TP53 wild type, I) Hey TP53 wild type, J) Kuramochi TP53 mutant, K) TykNu TP53 R175H mutant, and L) SKOV3 TP53 null cell lines. Asterisks indicate p < 0.05 by t-test. Error bars represent the SEM.
We treated ID8 murine ovarian cancer cell lines modified by CRISPR/Cas9 (ID8 Trp53+/+ and ID8 Trp53−/−) with Nutlin-3A to activate p53 and assessed transcription of murine REs. P21 (CDKN2A) was induced by Nutlin-3A in the Trp53+/+ but not the Trp53−/− cell line (Fig. S9A). Murine REs showed a similar pattern as in the human OC lines, as Nutlin-3A induced their expression in the Trp53+/+ but not Trp53−/− ID8 cells (Fig. S9B,C). Collectively, these data show that wild type p53 induces transcription of LTR loci in response to DNMTi treatment in both human and murine OC cells. This is consistent with the presence of p53 binding sites in many LTR elements (22).
The four OC cell lines profiled in Figures 1-6 differ in their TP53 mutation status and have other significant differences that include mutations in other tumor suppressors or oncogenes as well as cell doubling time. Thus we used CRISPR/Cas9 genome editing to introduce the R175H TP53 hotspot mutation into the TP53 WT Hey cell line (Fig. 7A,B, Fig. S10A-C). The R175H hotspot mutant Hey cell line exhibits baseline p53 protein expression, similar to the TykNu cell line (Fig. 6A, 7A). Nutlin-3A treatment induces expression of p53 target genes in the p53 WT Hey cell line but not the p53 R175H mutant cell line (Fig. 7B). We compared RE expression between the wild type (Hey WT, designated HC2) and mutant (Hey R175H, designated HH23) cell lines baseline and after DNMTi or Nutlin-3A (N3A) treatment. Baseline expression of RE families, including LTRs and SINEs (Fig. 7C) and individual loci (Fig. 7D) was greater in the Hey R175H compared to the Hey WT cell line. Upon Nutlin-3A activation of p53, only the WT cell line showed significant upregulation of REs, mostly LTRs (Fig. 7E,F). Upon DNMTi treatment, the Hey R175H cell line upregulated fewer REs compared to the Hey WT cell line (Fig. 7E,F). Combining DNMTi with Nutlin-3A p53 activation upregulated many more RE families and individual loci in the Hey WT cell line than either AZA or Nutlin-3A alone, but had minimal effects in the Hey R175H cell line (Fig. 7E,F). These data show that the hotspot R175H TP53 mutation causes an increase in RE expression baseline and that epigenetic therapy has less of an effect on RE transcription in the mutant line.
Discussion
Overall, these data demonstrate that epigenetic therapies upregulate diverse RE subfamilies and individual loci in OC cell lines and patient samples, and this upregulation is affected by TP53 status. HDACi alone had minimal effect on RE expression while DNMTi treatment significantly increased RE expression, notably at LTR, LINE, and SINE elements, and the combination of DNMTi/HDACi upregulated more REs than either treatment alone. These results are similar to regulation of genic sequences (49). Both LTR and SINE transcripts are protected from RNase digestion by MDA5 (17), and thereby transcripts from the RE subfamilies listed in Table S3 are potential triggers of interferon signaling and the subsequent anti-tumor response.
While there was significant variability in RE induction between cell lines, several RE subfamilies and individual transposable elements were upregulated in multiple cell lines following treatment with DNMTi and/or HDACi. Importantly, these REs were also found in OC patient samples. The RE subfamilies that are upregulated in multiple samples include HERV9-int and HERV-Fc1-int. HERV9 had previously been identified by Brocks et al. as commonly upregulated by DNMTi/HDACi in lung cancer cell lines (18) and we confirm this in OC cell lines. HERV9 elements are relatively recent integrations into the human genome are closely related to the HERVW elements that gave rise to Syncytin-1 (50). Syncytin-1 expression can occur due to loss of methylation in OC and pre-cancerous legions (51). SINEs, including Alu elements, were significantly demethylated and upregulated by DNMTi treatment in OC cell lines and showed less upregulation in TP53 mutant cell lines.
Mehdipour et al. (17) recently showed that DNMTi demethylation of orphan, often intronic CpG islands led to upregulation of inverted repeat Alu elements in colorectal cancer. The interferon signaling that resulted from the upregulation of Alu transcription decreased tumor size in treated mice (17). Their study found that the large majority of REs that bind to MDA5 and trigger immune signaling were Alus, though LTRs and other REs were also present. This is consistent with earlier work that showed Alu binding to MDA5 can be regulated by RNA editing (2,5). Since a growing body of evidence supports the role of Alus in the interferon response, it is interesting that we observe significant Alu demethylation by DNMTi in OC. We also found noticeable upregulation of Alus by DNMTi/HDACi treatment while DNMTi alone did not robustly upregulate these elements. This hints at a slightly different mechanism of action where histone acetylation may be more important for Alu regulation than in the Mehdipour et al. study, which could be caused by the differences between cancer types.
RE expression can be driven by non-epigenetic events including transcription factor binding. Our data show that p53 can transcriptionally activate REs in cancer cells. Hotspot TP53 mutants show higher baseline levels of RE expression and less RE upregulation upon DNMTi treatment (Fig. 5, 7). This may occur because TP53 mutant cell lines express p53 even in the absence of Nutlin-3A induction (Fig. 6A, 7A) and thus the p53 protein can bind and activate RE expression without treatment in TP53 mutant cell lines. Interestingly, combining DNMTi (AZA) treatment with Nutlin-3A induction of p53 induced many more RE families and individual loci than either treatment alone, but only in the p53 WT cell line (Fig. 7). This confirms transcriptional activation of REs by wild type p53, which is further enhanced by loss of methylation genome-wide. It is also interesting to note that HDACi can downregulate p53 expression (52). Though we did not examine the effects of ITF treatment on p53 expression in our cell lines, perhaps this effect contributes to the low levels of RE upregulation with ITF treatment alone. Thus, p53 plays a crucial role in regulation of RE transcription in OC. This role may be direct, as we infer at loci that exhibit p53 binding by ChIP-Seq (Fig. 4F), or indirect, through downstream signaling after p53 activation. p53 transcriptionally enhances interferon signaling (53) and recent work has shown that type I interferon signaling can induce LTR expression (54). When used at high doses, the DNMTi, AZA, can induce DNA damage and p53 activation, but at the low dose used in these experiments (500 nM) does not induce dsDNA breaks (55). Further mechanistic work is thus clearly needed to determine how wild type and mutant p53 regulate RE expression in the context of epigenetic therapies.
Data from primary OCs also correlates the presence of hotspot mutant p53 with RE expression. In ovarian cancers, STIC lesions—the carcinoma in situ precursors to high grade serous ovarian cancer—are characterized by mutant TP53 along with demethylation and increased expression of L1 elements (56). LINE-1 retrotransposition is limited by the p53 DNA damage response as well as interferon signaling and p53 directly represses human LINE1 (25,56). We observed p53 binding to LINE1 elements after Nutlin-3A or DNMTi treatment (Fig. 4F) but did not observe significant downregulation of LINE1 elements after these treatments (Fig. 7E,F). Instead, we observe p53 enrichment at and upregulation of the LTR and SINE RE classes (Fig. 4F, 6, 7). The ChIP-seq peaks we observed at REs were often at or near LTR10 elements (Fig. S11A-C). This is consistent with previous work showing that LTR10 elements contain p53 binding sites (22).
Limitations of our study include the etiology of the OC cell lines used. We recognize that the A2780 and Hey cell lines likely do not model high grade serous OC, while Kuramochi and TykNu are much better models of high grade serous OC (57,58) A2780 and Hey exhibit wild type TP53 and mutations found in other OC subtypes, including ARID1A. Differences in histological subtype between the four OC lines profiled likely contributes to the significant variability we observe in which RE families and individual loci are induced by DNMTi/HDACi treatment. As high grade serous OC is characterized by nearly 100% TP53 mutations (59), there is no TP53 wild type high grade serous cell line that we can use for our studies. To isolate the effects of p53, we thus generated isogenic TP53 wild type and mutant cell lines and analyzed the effects of epigenetic therapies and p53 activation in this controlled setting, showing a significant effect of p53 status on RE baseline transcription and upregulation by DNMTi (Fig. 7).
The importance of determining how and which REs are regulated by epigenetic factors and p53 is emphasized by ongoing clinical trials for combined epigenetic and immune therapy. A Phase Ib trial combining DNMTi treatment with anti-CTLA-4 to fight melanoma showed promising results, including improved immune activation and anti-tumor activity (60). This combination therapy is currently being tested in clinical trials for melanoma, colorectal cancer, ovarian cancer, and kidney cancer, among others (60) (NCT01928576, NCT02961101, NCT03019003, NCT02811497, NCT02546986, NCT02397720, NCT02530463). A recent study on TP53 hotspot mutations in acute myeloid leukemia showed that cancers with mutant TP53 exhibited higher interferon signaling, more infiltrating immune cells, and stronger response to immunotherapy, emphasizing the need for further study of how this transcription factor cooperates with epigenetic regulators to shape RE transcription and subsequent interferon signaling (61). Lastly, REs including ERVs are being actively investigated as potential cancer-specific antigen targets for immunotherapy (6,62-65). Understanding how the most commonly mutated protein in cancer affects RE expression, both baseline and with epigenetic therapy, will impact future immunotherapies for OC and other solid tumors.
Supplementary Material
Significance.
This study identifies the repetitive element targets of epigenetic therapies in ovarian cancer and indicates a role for p53 in this process.
Acknowledgements
We would like to acknowledge the Wang Lab for their assistance in training on methylMnM and methylCRF and their kind provision of scripts to update our annotation files to hg38. We would like to acknowledge Dr. Keith Crandall and Dr. Matthew Bendall for their assistance using the Telescope pipeline. We acknowledge the GW Pegasus Performance Computing core that manages the server on which we can store data and run analyses. We also acknowledge Evan Tarbell and Dr. Tao Liu for granting us early access to their tool HMMRATAC and for their aid as we established the pipeline in our lab. The ID8 Trp53−/− cells and ID8 CRISPR control cells were a kind gift from Dr. Iain McNeish, Imperial College London. We thank Castle Raley and the George Washington University Genomics Core for providing library construction and sequencing services. Research reported in this publication was supported by the National Cancer Institute under Awards R00CA204592 (to KBC), R21CA227259 (to KBC with RS as MPI), and R37CA251270 (to KBC) as well as by The Marlene and Michael Berman Endowed Fund for Ovarian Cancer Research. JIM was supported by an NCI NRSA Institutional Research Training Program grant (T32 CA 247756). SG was supported by an NRSA Predoctoral Fellowship (NIH/NCI 1F31CA254315-01). The authors would like to acknowledge the Institute for Biomedical Sciences at the George Washington University for graduate student support and training.
References
- 1.Kassiotis G, Stoye JP. Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol. 2016;16:207. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, et al. Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation. Cell. 2018;172:797–810.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med. 2018;24:1143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, et al. Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell. 2018;172:811–824.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong Y, Rose CM, Cass AA, Williams AG, Darwish M, Lianoglou S, et al. Transposable element expression in tumors is associated with immune infiltration and increased antigenicity. Nat Commun. 2019;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet. 2019;51:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodier JL. Restricting retrotransposons: a review. Mob DNA [Internet]. 2016. [cited 2017 Jan 30];7. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4982230/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41. [DOI] [PubMed] [Google Scholar]
- 10.Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh L-S, Lee S-L, et al. Approval Summary: Azacitidine for Treatment of Myelodysplastic Syndrome Subtypes. Clin Cancer Res. 2005;11:3604–8. [DOI] [PubMed] [Google Scholar]
- 11.Center for Drug Evaluation and Research. FDA approves Onureg (azacitidine tablets) for acute myeloid leukemia. FDA [Internet]. 2020. [cited 2020 Sep 24]; Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-onureg-azacitidine-tablets-acute-myeloid-leukemia [Google Scholar]
- 12.Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen R-WC, Vatapalli R, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, et al. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc Natl Acad Sci U S A. 1999;96:14007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karpf AR, Lasek AW, Ririe TO, Hanks AN, Grossman D, Jones DA. Limited Gene Activation in Tumor and Normal Epithelial Cells Treated with the DNA Methyltransferase Inhibitor 5-Aza-2′-deoxycytidine. Mol Pharmacol. 2004;65:18–27. [DOI] [PubMed] [Google Scholar]
- 15.Roulois D, Yau HL, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siebenkäs C, Chiappinelli KB, Guzzetta AA, Sharma A, Jeschke J, Vatapalli R, et al. Inhibiting DNA methylation activates cancer testis antigens and expression of the antigen processing and presentation machinery in colon and ovarian cancer cells. PLoS ONE. 2017;12:e0179501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehdipour P, Marhon SA, Ettayebi I, Chakravarthy A, Hosseini A, Wang Y, et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature. 2020;1–5. [DOI] [PubMed] [Google Scholar]
- 18.Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, et al. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet. 2017;49:1052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu M, Thomas SL, DeWitt AK, Zhou W, Madaj ZB, Ohtani H, et al. Dual Inhibition of DNA and Histone Methyltransferases Increases Viral Mimicry in Ovarian Cancer Cells. Cancer Res. 2018;78:5754–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walton J, Blagih J, Ennis D, Leung E, Dowson S, Farquharson M, et al. CRISPR/Cas9-Mediated Trp53 and Brca2 Knockout to Generate Improved Murine Models of Ovarian High-Grade Serous Carcinoma. Cancer Res. 2016;76:6118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone ML, Chiappinelli KB, Li H, Murphy LM, Travers ME, Topper MJ, et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc Natl Acad Sci. 2017;201712514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci. 2007;104:18613–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wylie A, Jones AE, D’Brot A, Lu W-J, Kurtz P, Moran JV, et al. p53 genes function to restrain mobile elements. Genes Dev. 2016;30:64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu W-J, Chapo J, Roig I, Abrams JM. Meiotic recombination provokes functional activation of the p53 regulatory network. Science. 2010;328:1278–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiwari B, Jones AE, Caillet CJ, Das S, Royer SK, Abrams JM. p53 directly represses human LINE1 transposons. Genes Dev. 2020;34:1439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018;25:154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Cancer Genome Atlas. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Topper MJ, Vaz M, Chiappinelli KB, Shields CED, Niknafs N, Yen R-WC, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell. 2017;171:1284–1300.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, et al. Analyzing CRISPR genome editing experiments with CRISPResso. Nat Biotechnol. 2016;34:695–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing X, Zhang B, Li D, Wang T. Comprehensive Whole DNA Methylome Analysis by Integrating MeDIP-seq and MRE-seq. DNA Methylation Protoc Methods Mol Biol Vol 1708 [Internet]. Humana Press, New York, NY; 2018. [cited 2017 Dec 22]. page 209–46. Available from: 10.1007/978-1-4939-7481-8_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2013-2015 [Internet]. Available from: http://www.repeatmasker.org [Google Scholar]
- 33.Jin Y, Tam OH, Paniagua E, Hammell M. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics. 2015;31:3593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendall ML, Mulder M de, Iñiguez LP, Lecanda-Sánchez A, Pérez-Losada M, Ostrowski MA, et al. Telescope: Characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLOS Comput Biol. 2019;15:e1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarbell ED, Liu T. HMMRATAC: a Hidden Markov ModeleR for ATAC-seq. Nucleic Acids Res. 2019;47:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M, Zhao Y, Zhang B. Efficient Test and Visualization of Multi-Set Intersections. Sci Rep. 2015;5:16923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gel B, Díez-Villanueva A, Serra E, Buschbeck M, Peinado MA, Malinverni R. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics. 2016;32:289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang F, Cardenas H, Huang H, Jiang G, Perkins SM, Zhang C, et al. Genomic and Epigenomic Signatures in Ovarian Cancer Associated with Resensitization to Platinum Drugs. Cancer Res. 2018;78:631–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moufarrij S, Srivastava A, Gomez S, Hadley M, Palmer E, Austin PT, et al. Combining DNMT and HDAC6 inhibitors increases anti-tumor immune signaling and decreases tumor burden in ovarian cancer. Sci Rep. 2020;10:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steiner MC, Marston JL, Iñiguez LP, Bendall ML, Chiappinelli KB, Nixon DF, et al. Locus-Specific Characterization of Human Endogenous Retrovirus Expression in Prostate, Breast, and Colon Cancers. Cancer Res. 2021;81:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanderKraats ND, Hiken JF, Decker KF, Edwards JR. Discovering high-resolution patterns of differential DNA methylation that correlate with gene expression changes. Nucleic Acids Res. 2013;41:6816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chuang JC, Warner SL, Vollmer D, Vankayalapati H, Redkar S, Bearss DJ, et al. S110, a 5-Aza-2′-Deoxycytidine–Containing Dinucleotide, Is an Effective DNA Methylation Inhibitor In vivo and Can Reduce Tumor Growth. Mol Cancer Ther. 2010;9:1443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine AJ, Ting DT, Greenbaum BD. P53 and the defenses against genome instability caused by transposons and repetitive elements. BioEssays. 2016;38:508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science. 2004;303:844–8. [DOI] [PubMed] [Google Scholar]
- 47.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leroy B, Girard L, Hollestelle A, Minna JD, Gazdar AF, Soussi T. Analysis of TP53 Mutation Status in Human Cancer Cell Lines: A Reassessment. Hum Mutat. 2014;35:756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. [DOI] [PubMed] [Google Scholar]
- 50.Vargiu L, Rodriguez-Tomé P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strissel PL, Ruebner M, Thiel F, Wachter D, Ekici AB, Wolf F, et al. Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: Emergence of new molecular targets. Oncotarget. 2012;3:1204–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan W, Liu S, Xu E, Zhang J, Zhang Y, Chen X, et al. Histone deacetylase inhibitors suppress mutant p53 transcription via histone deacetylase 8. Oncogene. 2013;32:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muñoz-Fontela C, Macip S, Martínez-Sobrido L, Brown L, Ashour J, García-Sastre A, et al. Transcriptional role of p53 in interferon-mediated antiviral immunity. J Exp Med. 2008;205:1929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsai H-C, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, et al. Transient Low Doses of DNA Demethylating Agents Exert Durable Anti-tumor Effects on Hematological and Epithelial Tumor Cells. Cancer Cell. 2012;21:430–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ardeljan D, Steranka JP, Liu C, Li Z, Taylor MS, Payer LM, et al. Cell fitness screens reveal a conflict between LINE-1 retrotransposition and DNA replication. Nat Struct Mol Biol. 2020;27:168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, et al. Type-Specific Cell Line Models for Type-Specific Ovarian Cancer Research. PLOS ONE. 2013;8:e72162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vang R, Levine DA, Soslow RA, Zaloudek C, Shih I-M, Kurman RJ. Molecular Alterations of TP53 are a Defining Feature of Ovarian High-Grade Serous Carcinoma: A Rereview of Cases Lacking TP53 Mutations in The Cancer Genome Atlas Ovarian Study. Int J Gynecol Pathol Off J Int Soc Gynecol Pathol. 2016;35:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giacomo AMD, Covre A, Finotello F, Rieder D, Danielli R, Sigalotti L, et al. Guadecitabine plus ipilimumab in unresectable melanoma: the NIBIT-M4 clinical trial. Clin Cancer Res. 2019;25:7351–62. [DOI] [PubMed] [Google Scholar]
- 61.Vadakekolathu J, Lai C, Reeder S, Church SE, Hood T, Lourdusamy A, et al. TP53 abnormalities correlate with immune infiltration and associate with response to flotetuzumab immunotherapy in AML. Blood Adv. 2020;4:5011–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cherkasova E, Scrivani C, Doh S, Weisman Q, Takahashi Y, Harashima N, et al. Detection of an Immunogenic HERV-E Envelope with Selective Expression in Clear Cell Kidney Cancer. Cancer Res. 2016;76:2177–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnamurthy J, Rabinovich BA, Mi T, Switzer KC, Olivares S, Maiti SN, et al. Genetic Engineering of T Cells to Target HERV-K, an Ancient Retrovirus on Melanoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21:3241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saini SK, Ørskov AD, Bjerregaard A-M, Unnikrishnan A, Holmberg-Thydén S, Borch A, et al. Human endogenous retroviruses form a reservoir of T cell targets in hematological cancers. Nat Commun. 2020;11:5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Natoli M, Gallon J, Lu H, Amgheib A, Pinato DJ, Mauri FA, et al. Transcriptional analysis of multiple ovarian cancer cohorts reveals prognostic and immunomodulatory consequences of ERV expression. J Immunother Cancer. 2021;9:e001519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The OC cell line RNA-seq, methylMnM, methylCRF, ChIP-seq, and ATAC-seq data are available in GEO, accession GSE182430. The trial data from the Fang et al. study (40) is available in GEO, accession GSE102120. TCGA data was accessed via the NIH GDC Data Portal (https://portal.gdc.cancer.gov/). Custom scripts from our laboratory used in the analyses presented here can be found on the Chiappinelli Lab Github page (https://github.com/Chiappinelli-Lab/Ovarian-Cancer-RE-Analysis-Scripts).