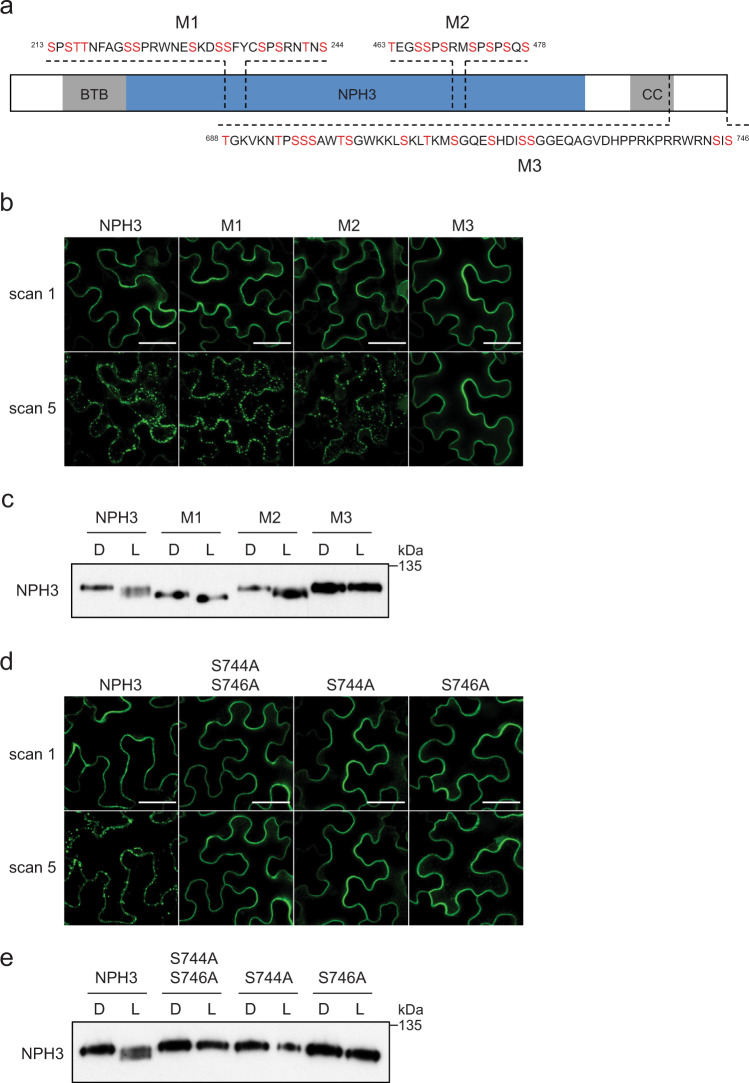
Fig. 2. Mutational analysis of NPH3 phosphorylation sites.
a Schematic illustration of the NPH3 protein indicating the location of the three mutagenized regions (M1, M2 and M3). For each region, all serine and threonine residues in red were substituted with alanine. The relative positions of the bric-a-brac, tramtrack and broad complex (BTB), NPH3 and coiled-coil (CC) domains are indicated. b Confocal images of GFP-NPH3 (NPH3) and phosphorylation site mutants (M1, M2 and M3) transiently expressed in leaves of N. benthamiana. Plants were dark-adapted before confocal observation and images acquired immediately (scan 1) and after repeat scanning with the 488 nm laser (scan 5). Bar, 50 µm. c Immunoblot analysis of protein extracts from leaves of N. benthamiana transiently expressing GFP-NPH3 (NPH3) and phosphorylation site mutants (M1, M2 and M3). Plants were dark-adapted and maintained in darkness (D) or irradiated with 20 µmol m−2 s−1 of blue light for 15 min (L). Protein extracts were probed with anti-GFP antibodies. d Confocal images of GFP-NPH3 (NPH3) and phosphorylation site mutants S744A S746A, S744A and S746A transiently expressed in leaves of N. benthamiana. Plants were dark-adapted before confocal observation and images acquired immediately (scan 1) and after repeat scanning with the 488 nm laser (scan 5). Bar, 50 µm. e Immunoblot analysis of protein extracts from leaves of N. benthamiana transiently expressing GFP-NPH3 (NPH3) and phosphorylation site mutants S744A S746A, S744A and S746A. Plants were dark-adapted and maintained in darkness (D) or irradiated with 20 µmol m−2 s−1 of blue light for 15 min (L). Protein extracts were probed with anti-GFP antibodies. Experiments were repeated at least twice with similar results.