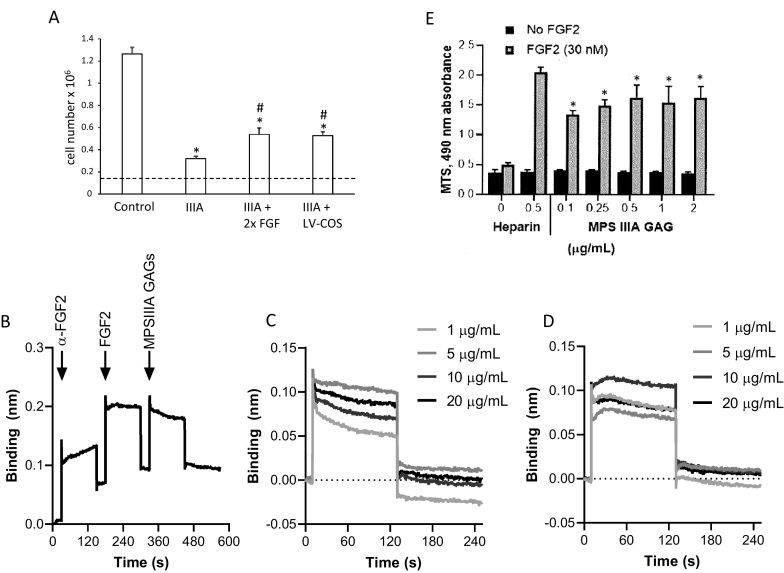
Fig. 3.
Rescue of iPSC-derived NPC proliferation and role of MPS IIIA GAG on FGF2 binding and signalling. (A) Control iPSC-derived NPCs, MPS IIIA-intermed iPSC-derived NPCs and MPS IIIA-intermed iPSC-derived NPCs transduced with a lentivirus encoding sulphamidase (LV-COS) were seeded at 150,000 cells per 24-well and maintained in neural expansion medium. Additional cultures of MPS IIIA-intermed iPSC-derived NPCs received a 2-fold increase in FGF2 in the culture medium (i.e., 40 ng/mL instead of 20 ng/mL). Cell number was determined 7 days after seeding and results expressed as the mean ± std. dev of n = 3 replicates. *denotes significant difference to control and # denotes significant difference between MPS IIIA-intermed iPSC-derived NPCs treated with additional FGF2 or transduced with a lentivirus encoding sulphamidase and MPS IIIA-intermed iPSC-derived NPCs (P < 0.05). (B) MPS IIIA GAG binding to FGF2 was determined using FGF2 immobilised to a protein G sensor via an anti-FGF2 antibody followed by the addition of FGF2 and then MPSIIIA GAGs as shown or heparin (not shown). (C) Binding curves for MPSIIIA GAG at concentrations between 1 and 20 μg/mL. (D) Binding curves for heparin at concentrations between 1 and 20 μg/mL. (E) Proliferation of BaF32 cells expressing FGFR1c in the presence of 0.03 nM FGF2 and either heparin or MPSIIIA GAG. Results are expressed as the mean ± std. dev of n = 3 replicates. * denotes significant difference to cells grown in the absence of FGF2 and ** denotes significant difference to cells grown in the presence of FGF2 and all concentrations of MPSIIIA GAGs (P < 0.05).