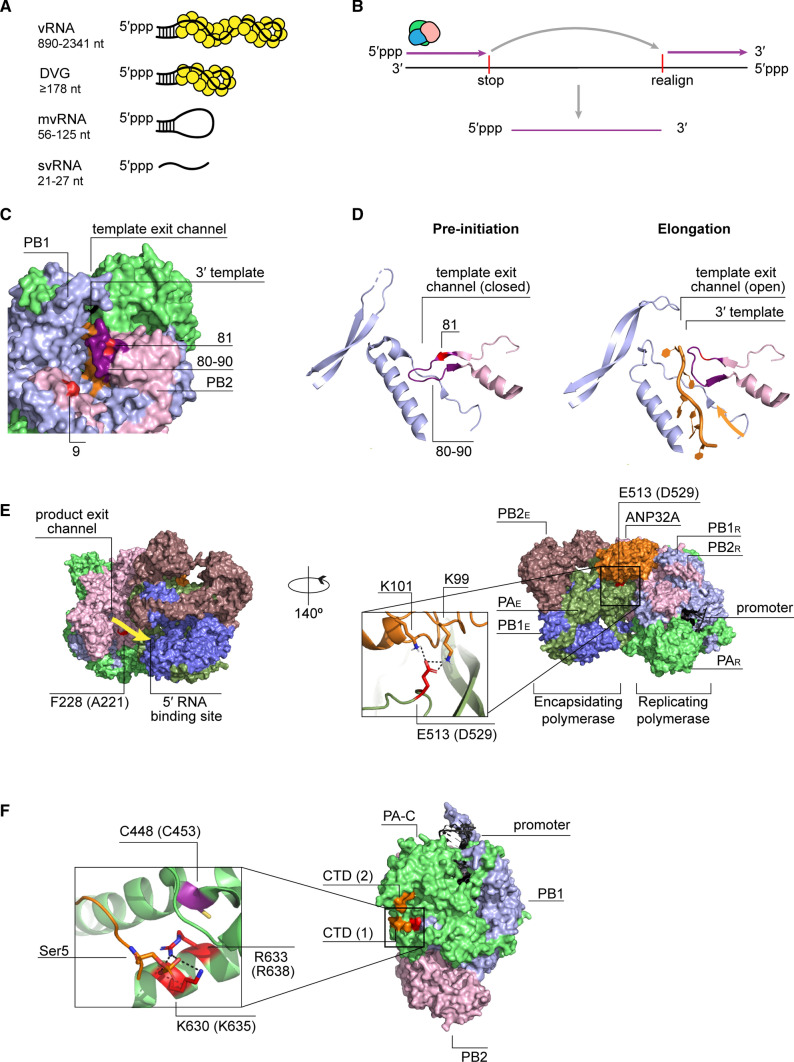
Fig. 4.
Aberrant viral RNA formation and RNA polymerase residues putatively implicated in this process. A A schematic representation of different types of aberrant RNAs produced by the influenza A virus polymerase. B A possible mechanism for the DVG and mvRNA synthesis during replication. C A surface representation of the bat H17N10 influenza A polymerase (PDB 6T0V) during transcription elongation. The 3′ terminus of the template (orange) can be seen exiting through the template exit channel. PB2 residues 9 and 81 are highlighted in red and PB2 80–90 loop is shown in purple. D A cartoon representation of the template exit channel of the bat H17N10 influenza A polymerase at the stage of transcription pre-initiation (PDB 6T0N) and during transcription elongation (PDB 6T0V). The 80–90 loop (purple) undergoes an outward movement, allowing opening of the template exit channel during elongation. Direction of the exiting template is indicated with an arrow. E A surface representation of the replicating-encapsidating polymerase dimer of the influenza C virus (ICV) polymerase in the complex with chicken ANP32A (PDB 6XZR). Residue numbering is as in ICV RNA polymerase. The corresponding IAV RNA polymerase residues are shown in parenthesis. PB2 residue F228 of ICV (A221 IAV) is located in an RNA path (yellow arrow) between the product exit channel of the replicating polymerase and the 5′ binding site of the encapsidated polymerase. Hydrogen bonds between the PA E513 of ICV (D529 in IAV) and two lysines of ANP32A (K99 and K101) are shown. F A surface representation of the bat H17N10 influenza A virus bound to the CTD of Pol II (PDB 5M3H). Two binding sites for Pol II CTD (orange) are shown. Residue numbering as in bat IAV, and corresponding human and avian residues are shown in parenthesis. PA residues, K630 and R633 of the bat IAV polymerase (corresponding to K635 and R638 of the human or avian IAV polymerase, respectively) are shown to form hydrogen bonds with phosphorylated Ser5 of the Pol II CTD. PA residue C448 in the bat IAV polymerase (C453 in the human or avian IAV) is indicated in purple