Fig. 6. Overview of flg22- and D-glucose-biased signaling.
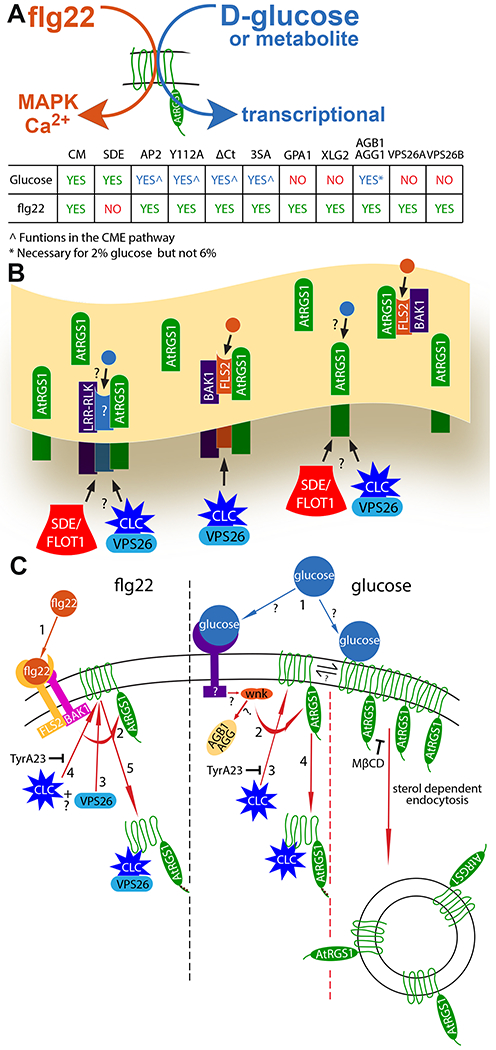
(A) Simple model illustrating the biased signaling output through AtRGS1 leading from flg22 and glucose or metabolite input, and a chart summarizing the origins of endocytosis (CME and SDE), recognition motif and phosphorylation requirements, and individual proteins necessary for glucose- and flg22-induced endocytosis of AtRGS1 where green “yes” indicates a requirement for signaling, red “no” indicates not required, and blue “yes” indicates a requirement with some restrictions. (B) Membrane overview illustrating proposed AtRGS1 microdomain clusters with common RLK neighbors. flg22 (orange circle) binds to FLS2 to initiate signaling through AtRGS1. The mechanism of glucose (blue circle) perception is unknown as indicated by the question marks. After ligand perception, SDE or CME of AtRGS1 occurs to permit downstream signaling. (C) A diagram of the individual components involved in the mechanism of endocytosis initiated by flg22 and glucose or metabolite and the presumed sequence of events. For flg22: (step 1) perception of ligand, (step 2) phosphorylation of AtRGS1 by a RLK, (steps 3 and 4) binding of clathrin complex and/or VPS26 in an unknown order, and (step 5) internalization of AtRGS1. For glucose or metabolite: (Step 1) ligand perception by a RLK or direct interaction with AtRGS1, (step 2) receptor interaction with WNK (for SDE, step 2 indicates immediate endocytosis because other key components of the pathway are unknown), (step 3) phosphorylation of AtRGS1 by WNK, (step 4) binding of clathrin complex to AtRGS1, and (step 5) internalization of AtRGS1. TyrA23 inhibits CME in both pathways, and MβCD inhibits AtRGS1 microdomain formation at the membrane.
