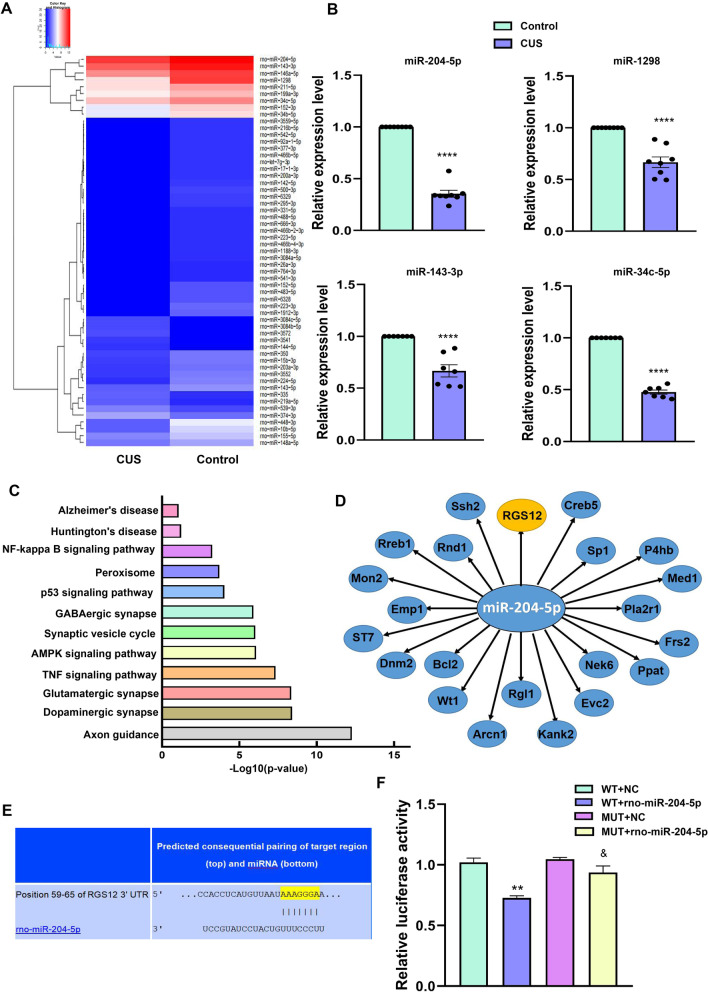
Fig. 2.
MiR-204-5p is reduced in the hippocampal DG region of CUS rats. A Heat map of differentially expressed miRNAs in hippocampal DG regions. B Relative expression levels of miR-204-5p, miR-1298, miR-143-3p and miR-34c-5p in the DG region (N = 7–8 per group). **P < 0.01, ****P < 0.0001, vs Control group. C Bar graphs represent magnitudes of significant correlations for miR-204-5p mediated signaling pathways as indicated by respective P-values (− log10 scaled). D Bioinformatical prediction of miR-204-5p target genes. E Predicted putative seed-matching sites between miR-204-5p and RGS12. F Luciferase reporter assay results as performed on 293Tcells to detect relative luciferase activities of WT and MUT RGS12 reporters (N = 3/group). **P < 0.01 vs WT + NC. &P < 0.05 vs WT + rno-miR-204-5p. Data are presented as means ± SEMs. Student t-tests were employed for comparisons between the two groups. One-way ANOVA followed by Tukey’s post-hoc test was used for multiple comparisons involving > 2 groups