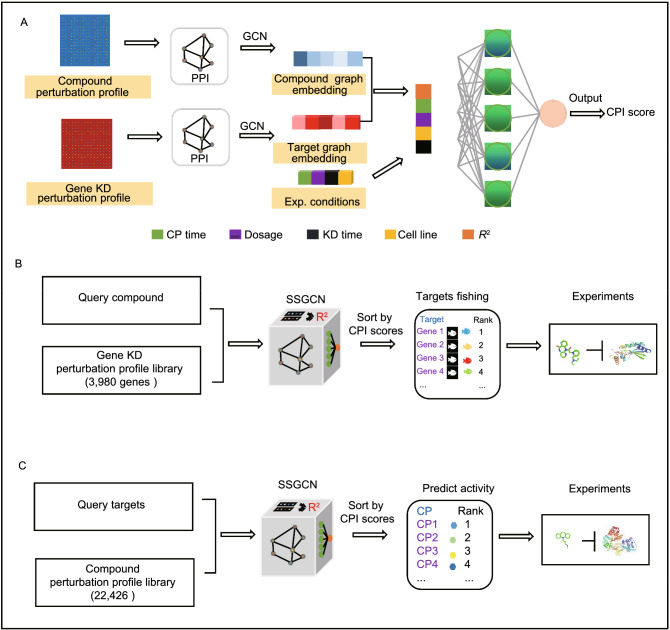
Figure 1.
Target prediction using the SSGCN model. (A) Architecture of the SSGCN. Compound graph embedding is obtained by a spectral-based graph convolutional network (GCN) to integrate the protein-protein interaction (PPI) network topological structure information and compound perturbation profile. Target graph embedding is obtained by another GCN to integrate PPI and gene knockdown perturbation profile. The correlation coefficient Pearson R2 is calculated between the compound graph embedding and target graph embedding. The CP time is the duration of compound (CP) treatment and the KD time is the duration of gene knockdown (KD) perturbation. CPI score is the classification probability of whether the compound interacts with the protein. (B) Pipeline of the target inference using the SSGCN model. (C) Pipeline of identifying the novel active compound using the SSGCN model