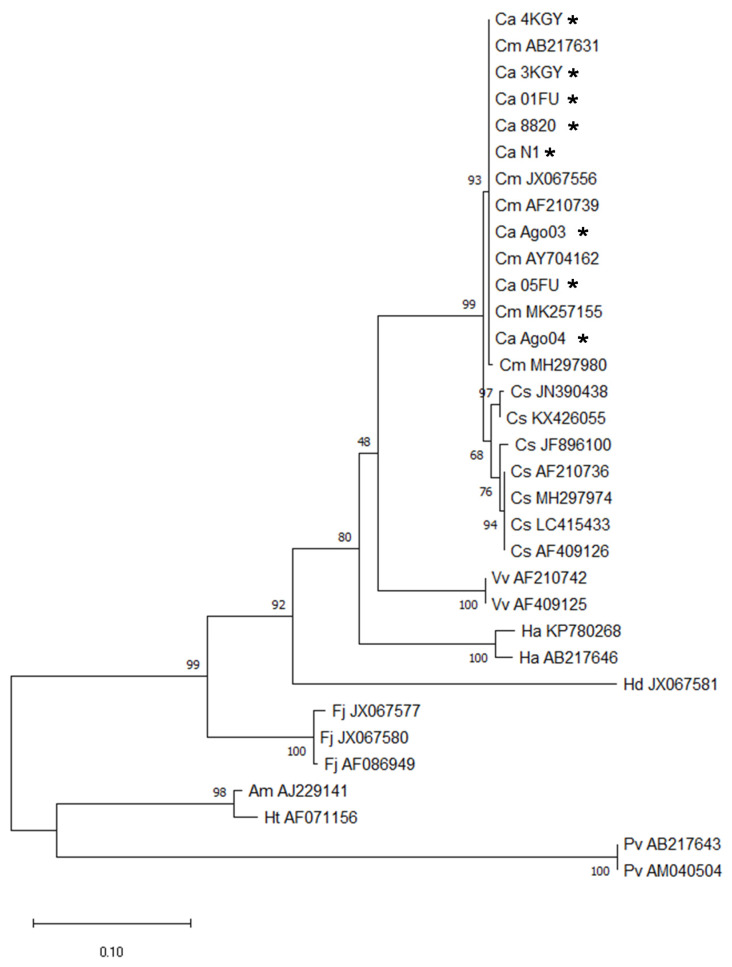
Figure 1.
Maximum-likelihood phylogenetic tree from partial sequences of the large subunit (LSU) D1–D2 regions in rDNA of Chattonella marina complex strains. The tree was inferred from the K2 + G model. The accession numbers or strain ID used in the present study (asterisks) are shown following the species name. Numbers on the major nodes present maximum-likelihood bootstrap values (1000 replicates). The tree was rooted using Ascoseira mirabilis, Halosiphon tomentosus, and Psuedochattonella verruculosa as the outgroup. Abbreviations of scientific names are as follows: Ca, Chattonella antiqua; Cm, C. marina; Cs, C. subsalsa; Vv, Vacuolaria virescens; Ha, Heterosigma akashiwo; Hd, Haramonas dimorpha; Fj, Fibrocapsa japonica; Am, A. mirabilis; Ht, H. tomentosus; Pv, P. verruculosa.