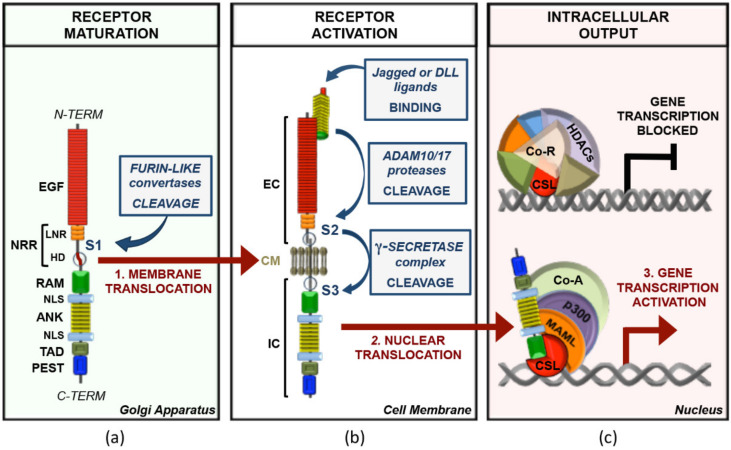
Figure 1.
Main framework of canonical Notch signal transduction. (a) Notch receptors are synthetized as monomeric precursor proteins, which are subjected to a proteolytic cleavage by furin-like convertases (S1) in the Golgi apparatus before being exposed to the cell membrane as non-covalently linked heterodimers. From the N- to the C-terminal, the mammalian Notch proteins comprises: EGF (epidermal growth factor-like repeats), NRR (negative regulatory region), LNR (Lin12/Notch repeats), HD (heterodimerization domain), RAM (RBP-jk associated molecule), NLS (nuclear localization signal), ANK (ankyrin repeats), TAD (transactivation domain), and PEST (proline, glutamic acid, serine, and threonine). (b) The interaction of a Jagged or DLL (Delta-like ligand) family ligand to the EC (extracellular fragment) of the trans-membrane Notch receptor leads to the S2 cleavage of the receptor by ADAM10/17 (a disintegrin and metalloproteinase) and the subsequent S3 proteolysis catalyzed by the γ-secretase complex. This last cleavage releases from the membrane the IC (intracellular fragment) of Notch, which translocates to the nucleus. (c) In the absence of Notch, the transcription factor CSL (CBF-1/SuH/Lag-1 DNA-binding protein), in association with several Co-R (co-repressors factors) and HDACs (histone deacetylases) on the regulatory regions of Notch target genes, acts as a transcriptional repressor. The binding of the Notch IC to CSL displaces from CSL the Co-R, and by recruiting MAML (Mastermind-like), p300, and distinct context-related Co-A (co-activators factors), target genes’ transcription is switched to an activated state.