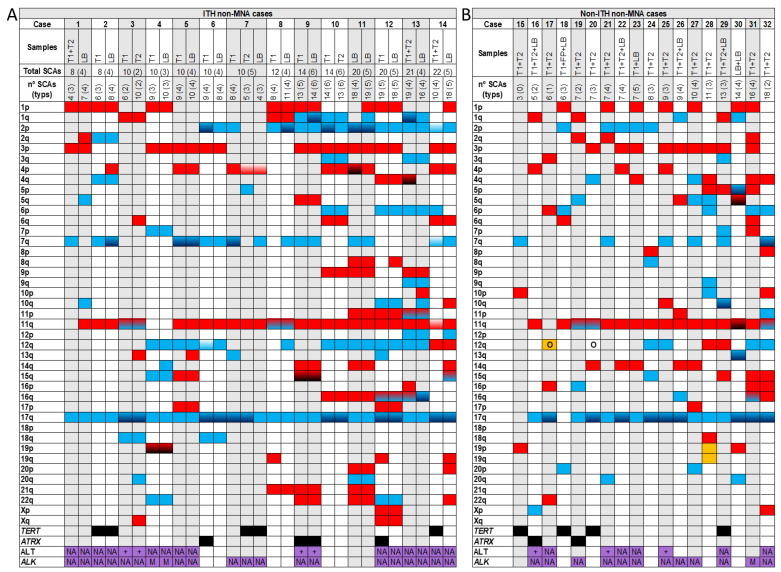
Figure 1.
Genomic aberrations found by SNPa in MYCN non amplified cases (non-MNA) with intra-tumour heterogeneity (ITH) (A) and non-ITH (B). In the upper part of both Figures A and B the analysed samples (T: solid tumour -1, 2-; LB: liquid biopsy) corresponding to each case (1–32) with the total number of segmental chromosomal aberrations (SCAs) and the typical ones in parenthesis, are shown. In Figure 1A the total number of SCAs and typical ones of each case, taking into account all the analysed samples, are also presented. Solid blue  or red
or red  squares refers to gains or losses, respectively in the indicated chromosome arm. Gradient color from blue
squares refers to gains or losses, respectively in the indicated chromosome arm. Gradient color from blue  or red
or red  to black indicates gains or losses with two or more chromosome fragments in the chromosome arm. Gradient color from white to blue
to black indicates gains or losses with two or more chromosome fragments in the chromosome arm. Gradient color from white to blue  or red
or red  indicates changes in the break position (which results in size variations of the aberration) in the chromosome arm between the analyzed samples of a case (only in ITH ones). Gradient color from blue to red
indicates changes in the break position (which results in size variations of the aberration) in the chromosome arm between the analyzed samples of a case (only in ITH ones). Gradient color from blue to red  refers to both gains and losses in the same chromosome arm of a sample. Solid yellow
refers to both gains and losses in the same chromosome arm of a sample. Solid yellow  squares indicate the presence of a chromotripsis-like phenomenon. Black squares
squares indicate the presence of a chromotripsis-like phenomenon. Black squares  indicate the presence of focal segmental chromosomal aberrations (FSCAs) affecting TERT or ATRX genes. Purple squares
indicate the presence of focal segmental chromosomal aberrations (FSCAs) affecting TERT or ATRX genes. Purple squares  indicate that the samples have been analysed for alternative lengthening of telomeres (ALT) or for mutations and amplifications in the ALK gene; NA: in the squares indicates that any aberration has been detected in the sample; +: refers to the presence of ALT; A: refers to presence of ALK amplification; M: indicates ALK mutation. O: indicates amplification of any gene or region in the chromosome arm, excluding MYCN or ALK amplification.
indicate that the samples have been analysed for alternative lengthening of telomeres (ALT) or for mutations and amplifications in the ALK gene; NA: in the squares indicates that any aberration has been detected in the sample; +: refers to the presence of ALT; A: refers to presence of ALK amplification; M: indicates ALK mutation. O: indicates amplification of any gene or region in the chromosome arm, excluding MYCN or ALK amplification.