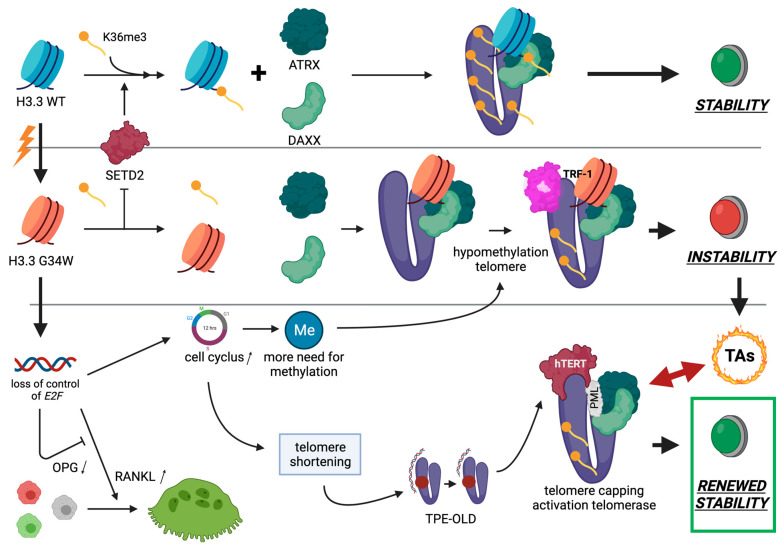
Figure 3.
Cell biology of GCTB. See left upper corner. Mutation leading from wild-type H3.3 (blue) to H3.3G34W (orange) inhibits the activity of the histone methyltransferase SETD2, which is in normal circumstances responsible for H3K36me3 of the histone tail. Wild-type H3.3 telomeres (purple) are sufficient methylated, and form together with ATRX and DAXX stable telomere ends. In contrast, affecting the H3.3-G34W terminal tail leads to a relative hypomethylation status. The loss of the telomeric H3K36me mark could favour the binding of TRF-1 to telomeres resulting in a relative telomeric instability further leading to mainly sister–sister TAs. In H3.3G34W mutated stromal cells E2F targets are the most downregulated genes. This suggests that the high proliferative activity of these mutated cells may be the result of silencing negative regulators of the cell cycle, most probably the E2F4-6 family members. When taking the GCTB-relevant genes targeted by the E2F transcription factors into account, RANKL and osteoprotegrin (OPG) are differentially targeted which leads to a loss of E2F control resulting in secretion of high levels of RANKL together with reduced levels of its decoy receptor OPG, and therefore to an enhanced osteoclastogenesis. Due to a higher proliferative activity there is more need for methylation of the telomeres, while these hypomethylated telomeres shorten faster. As a result of this shortening, the silencing loop structure of TPE-OLD is no longer in the proximity of hTERT, leading to hTERT transcription and telomerase activity. Together with the formation of a structural telomere protective cap (PML/ATRX/DAXX) this whole mechanism plays a pivotal role in successful and stable telomere maintenance leading to a renewed telomeric and hereby genomic stability.