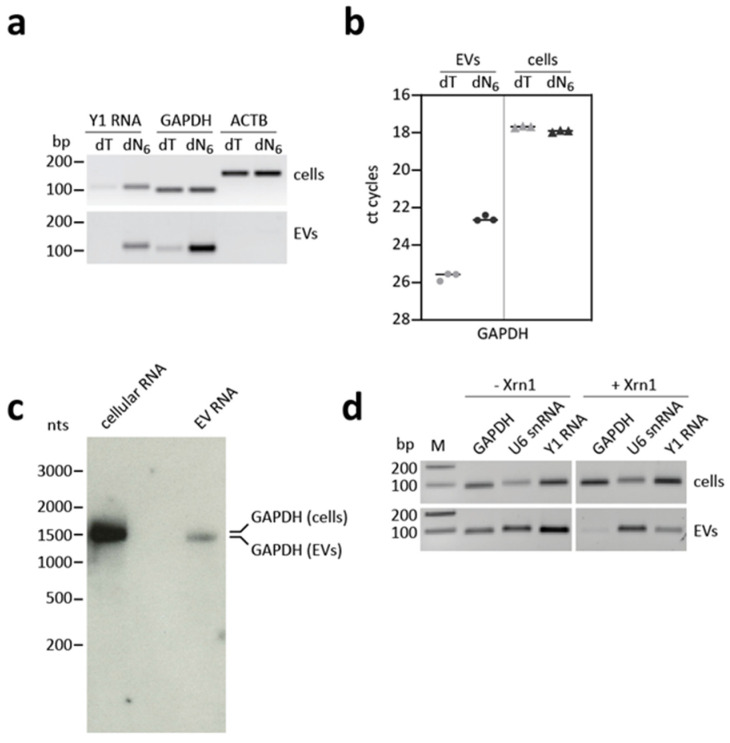
Figure 4.
EV-associated and cellular GAPDH mRNA differ in size and polyadenylation status. (a) Total RNA from ES-2 cells and their corresponding EVs were isolated, followed by RT-PCR. For reverse transcription, either oligo(dT) or random hexamers (dT, dN6) were used. Markers (200, 100 bp). (b) RNA was isolated from ES-2 cells and their corresponding EVs. For reverse transcription, either oligo(dT) or random hexamers (dT, dN6) were used, and GAPDH mRNA was quantified by RT-qPCR. (c) Detection of GAPDH mRNA in EVs by the glyoxal Northern blot analysis. Total cellular and EV-RNA (300 ng) were analyzed by glyoxal agarose gel electrophoresis and Northern blotting. Exposure time 3 min (d) Total RNA from ES-2-derived EVs was isolated and treated with Xrn1 exoribonuclease (+Xrn1). As control, no enzyme was added (−Xrn1). RT-PCR was performed using primers directed against transcripts indicated. Markers (200, 100 bp).