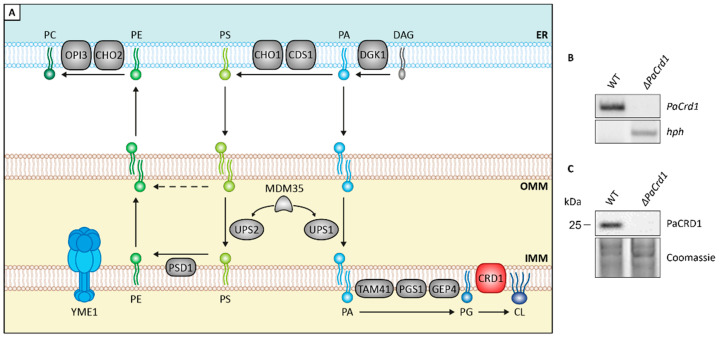
Figure 2.
Genetic modulation of PL homeostasis. (A): Scheme depicting major components of PL biosynthesis pathways in yeast. ER: endoplasmatic reticulum; OMM: outer mitochondrial membrane; IMM: inner mitochondrial membrane; CDS1: PA cytidylyltransferase; CHO1: PS synthase; CHO2: PE methyltransferase; CRD1: CL synthase; DGK1: DAG kinase; GEP4: phosphatidylglycerophosphatase; OPI3: PL methyltransferase; PGS1: phosphatidylglycerophosphate synthase; PSD1: PS decarboxylase proenzyme 1; TAM41: PA cytidylyltransferase; UPS1: unprocessed MGM1 protein 1; UPS2: unprocessed MGM1 protein 2; YME1: yeast mitochondrial escape protein 1. For other abbreviations, see caption of Figure 1. (B): Southern blot analysis with HindIII-digested DNA of P. anserina wild-type (WT) and ΔPaCrd1 strains demonstrating replacement of the PaCrd1 ORF with a hygromycin resistance gene (hph) in the deletion mutant. (C): Western blot analysis of PaCRD1 levels in isolated mitochondria of WT and ΔPaCrd1. Coomassie was used as loading control.