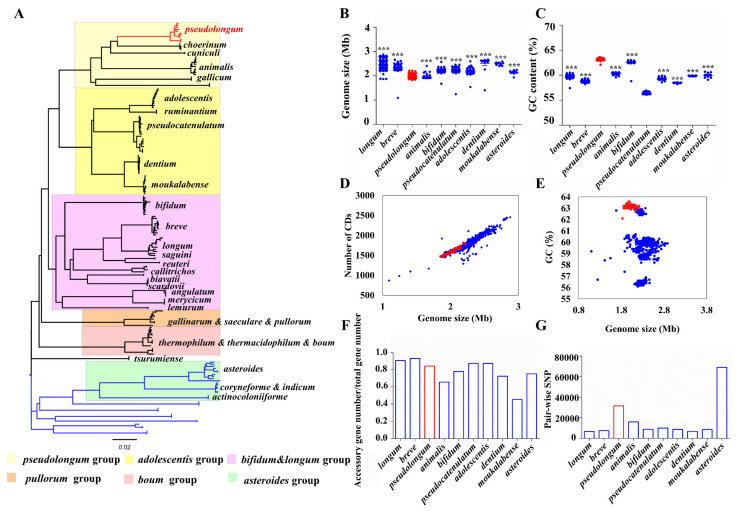
Figure 4.
Features of B. pseudolongum in terms of phylogenetic role and general genomic features among various Bifidobacterium species. (A) Phylogenetic tree (NJ tree) of genus Bifidobacterium based on the protein sequences of nine orthologs of various species. For species with more than 10 publicly available sequenced genomes, 10 representative strains distributed across the tree, which covered the overall genetic distance of each species, were selected; for those with less than 10 publicly available sequenced genomes, all the strains were included. (B,C) Genome size (Mb) (B) and GC content (%) (C) of different species. The statistical analysis was conducted between B. pseudolongum and each of the other species. ***, p < 0.001. (D) Association between genome size (Mb) and the number of CDSs, Pearson r = 0.94, p < 0.0001. (E) Association between genome size (Mb) and GC content (%), Pearson r = −0.3, p < 0.0001. (F,G) Comparison of intra-species genomic diversity for each species by accessory genome size (F) and SNP distance (G). Columns and dots representing B. pseudolongum are highlighted in red. All the available 887 sequenced genomes of Bifidobacterium in the NCBI database were used for analysis (A). Each of the Bifidobacterium species with more than 10 publicly available sequenced genomes in the NCBI database were analyzed, resulting in 786 strains in total (B–G).