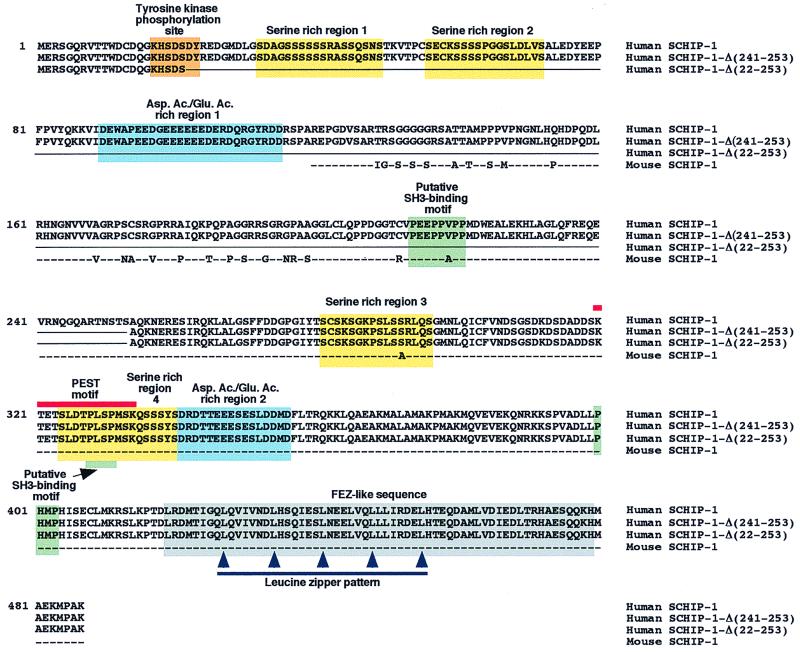
FIG. 1.
Protein sequence and putative domain structure of SCHIP-1. Line 1, amino acid sequence of human SCHIP-1, deduced from the nucleotide sequence of a SCHIP-1 cDNA, isolated from a λgt10 human fetal brain library. Lines 2 and 3, amino acid sequences of two putative isoforms of human SCHIP-1, named SCHIP-1-Δ(241-253) and SCHIP-1-Δ(22-253). The corresponding cDNAs were identified by reverse transcriptase PCR amplification experiments on human brain poly(A)+ RNA. These cDNAs may correspond to in-frame alternative splicing events. Amino acids absent in SCHIP-1-Δ(241-253) and SCHIP-1-Δ(22-253) are replaced by a continuous line. Line 4, amino acid sequence encoded by the partial mouse SCHIP-1 cDNA isolated from the two-hybrid screen. Dashes mark identity between human and mouse sequences; nonconserved amino acids between human and mouse proteins are directly indicated. Yellow and blue boxes indicate the four serine-rich regions and the two aspartic acid/glutamic acid-rich regions of the human SCHIP-1 protein, respectively. Green boxes show the localization of three putative SH3-binding domains conserved between the mouse and human SCHIP-1 proteins, and the orange box shows the localization of a putative tyrosine kinase phosphorylation site in human SCHIP-1. The grey box indicates the region of homology between the SCHIP-1 and FEZ proteins, predicted to adopt a coiled-coil conformation. The red line defines a putative PEST motif conserved in mouse and human SCHIP-1 proteins, and the blue line indicates the leucine zipper pattern in the putative coiled-coil region.